Abstract
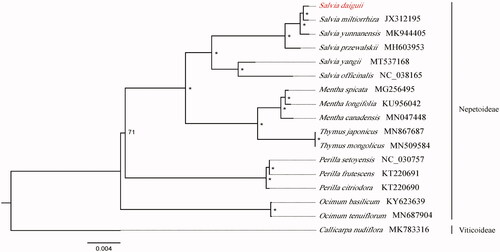
The complete chloroplast genome of Zhangjiajie sage, Salvia daiguii, was assembled in this study. The genome is 151,434 bp in length and contained 134 encoded genes in total, including 88 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. The result of phylogenetic analysis based on 17 chloroplast genomes revealed that S. daiguii is clustered with Salvia miltiorrhiza in Lamiaceae.
Zhangjiajie sage, Salvia daiguii Y. K. Wei & Y. B. Huang, is a newly described species in Salvia subgenus Glutinaria (Wei et al. Citation2019). The species has a unique streamside habitat with limestone bedrock, and is only known from Wulingyuan area, Zhangjiajie National Forest Park, Hunan province, China. The discovered population comprises no more than four hundred mature individuals and the number show dramatic fluctuations from year to year. S. daiguii has been classified as ‘Critically Endangered’ (CR) according to IUCN guidelines and suggest that immediate measures should be taken to ensure the long-term survival of this species in its original habitat (Wei et al. Citation2019). It usually grows in a compact form with sub-leathery and glossy leaves, and inflorescence is prominent. Therefore, S. daiguii has potential value of ornamental horticulture. Additionally, recent study showed that root of S. daiguii has high composition of caffeic acid, salvianolic acid B, and rosmarinic acid (Wang Citation2020). However, no genetic resource has been reported for this CR species. In this study, the complete chloroplast genome of S. daiguii was assembled and annotated, which will provide organelle molecular basis for this species and enlarge the genetic resources of genus Salvia.
The S. daiguii individual was collected from Zhangjiajie, Hunan, China (GPS: 29°2′7.60′′N, 110°29′12.19′′E, voucher S0297, deposited at Herbarium of Shanghai Chenshan Botanical Garden (CSH)). DNA was extracted from its leaf using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA). The raw data were generated using the Illumina platform (Illumina Inc., San Diego, CA). In total, about 3.8 G high-quality clean reads (150 bp PE read length) were obtained with adaptors trimmed. Following Liu et al. (Citation2017, Citation2018), the complete chloroplast genome was assembled with rbcL gene as seed using software Novoplasty (Dierckxsens et al. Citation2017). Aligning and annotation were conducted by GeSeq (Tillich et al. Citation2017) and Geneious Prime (Biomatters Ltd, Auckland, New Zealand).
The complete chloroplast genome of S. daiguii (GenBank accession No. MT901216) has a length of 151,434 bp with a typical circle structure and a GC content of 38%. It consists of a large single-copy region (LSC, 82,753 bp, 36.1% GC content), a small single-copy region (SSC, 17,576 bp, 32.1% GC content), and two inverted repeat regions (IR, 25,553 bp, 43.1% GC content). In total, there were 134 genes in S. daiguii, including 88 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Among them, six protein-coding genes (rpl2, rpl23, ycf2, ycf15, ndhB, and rps7), seven tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU) and all four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5) have two copies. Nine of these protein-coding genes (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, and ndhA) have one intron each and two (ycf3 and clpP) of them have two introns.
Seventeen species with available chloroplast genomes in Lamiaceae were selected to study the phylogenetic placement of S. daiguii (). The sequence alignment was conducted by MAFFT version 1.3 (Katoh and Standley Citation2013). We drew phylogenetic tree by the software IQ-TREE (Nguyen et al. Citation2015) with 5000 bootstrap replicates and TVM + F+R2 model. The result of phylogenetics analysis showed that S. daiguii is closely related to S. miltiorrhiza.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Raw sequencing data have been deposited in SRA (SRR12667953). The DNA matrix and phylogenetic tree that support the findings of this study are openly available in GitHub at https://github.com/andresqi/Zhejiangjie-sage-chloroplast-genome.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu LX, Li P, Zhang HW, Worth JRP. 2018. Whole chloroplast genome sequences of the Japanese hemlocks, Tsuga diversifolia and T. sieboldii, and development of chloroplast microsatellite markers applicable to East Asian Tsuga. J Forest Res. 23(5):318–323.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese Bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:15.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang FY. 2020. Diversity of chemical constituents of Salvia species in China. Hangzhou (China): Zhejiang Sci-Tech University.
- Wei YK, Pendry CA, Zhang DG, Huang YB. 2019. Salvia daiguii (Lamiaceae): a new species from west Hunan, China. Edinb J Bot. 76(3):359–368.