Abstract
The complete mitochondrial genome of the sun loach (Yasuhikotakia eos) was determined based on Illumina data in this study. The result showed that the closed double-stranded circular mitogenome was 16,738 bp in total length (GenBank accession number: MT800510) with 58.41% AT. The mitochondrial DNA consisted of 13 protein-coding genes (PGCs), 22 transfer RNA (tRNA) genes, 2 ribosomal (rRNA) genes, and 1 non-coding control region. Phylogenetic analysis suggested that Y. eos was most closely related to its congener Y. modesta. This work provides molecular information for further research on species identification and evolutionary relationships.
The sun loach (Yasuhikotakia eos), belonging to the subfamily Botiinae (Cypriniformes: Cobitidae), was known to be mainly distributed in the Mekong River, Chao Phraya, and Maeklong basins in Asia (Rainboth Citation1996). This species is extremely aggressive as other belligerent loach species and inhabits in rapidly flowing rivers. Due to the similar morphology, Y. eos was easily confused with its sister species during the taxonomic identification, particularly for the smaller or incomplete specimen (Nalbant Citation2002). Here, we described the characterization of the mitochondrial genome of Y. eos and explored the phylogenetic relationship among Botiinae species, which were expected to contribute to the further studies on taxonomic resolution and phylogenetic relationships of these taxa.
Yasuhikotakia eos was collected in Mekong River (12°46′33.2′′N, 105°57′29.6′′E), Cambodia in December 2019, and were deposited in Mekong Fish Herbarium of Cambodian First Unite Construction Group (Specimen ID: MK2019040). Tiny tail fin tissues were obtained and preserved in 95% ethanol. Total genomic DNA was extracted with a QIAamp DNA mini kit (Qiagen, Germany) following the manufacturer’ instructions. After monitored, the genomic DNA was sequenced through Illumina Hiseq 2500 platform (Illumina, San Diego, CA), and the mitogenome was assembled by metaSPAdes software (Nurk et al. Citation2017) using Yasuhikotakia morleti as reference.
The complete mitochondrial genome of Y. eos was a circular molecule with 16,738 bp in total length (GenBank accession number: MT800510). The overall nucleotides composition was 32.34%, 25.98%, 26.31%, and 15.28% for A, T, C, and G respectively, which showed a bias toward A and T (58.41%). Similar to the typical mitogenome of vertebrates, Y. eos also contained a set of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 non-coding control region (Supplementary Figure S1) (Ma et al. Citation2014; Wang et al. Citation2019). Among these 37 genes, 9 genes (ND6, tRNACys, tRNATyr, tRNASer, tRNAAla, tRNAGln, tRNAGlu, tRNAAsn, and tRNAPro) were encoded on the light (−) strand, while the remaining 28 genes were located at the heavy (+) strand. Most of the PCGs used ATG as the initiation codon except ND5 (started with ATA), COI (started with GTG), and ND3 (started with ATT). Besides, 10 PCGs terminated with TAA codon, but ND2, ND3, and ND4 used TAG as the stop codon. The length of 22 tRNAs ranged from 66 bp (tRNACys) to 76 bp (tRNALys). Two rRNA genes, 12S RNA (950 bp) and 16S RNA (1673 bp) were distributed between tRNAPhe and tRNALeu, separated by tRNAVal. The D-loop region was located between tRNAPhe and TrnaPro with 1080 bp in length.
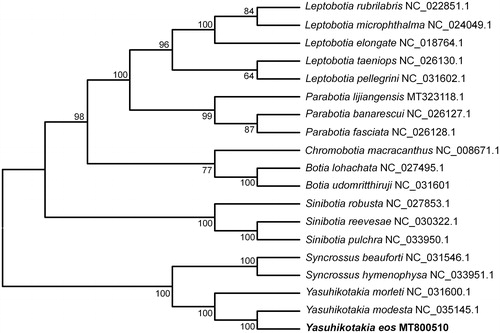
The mitochondrial genes have been widely used for species identification and inferring phylogenetic relationships so far (Frezal and Leblois Citation2008; Kochzius et al. Citation2010). Based on the 13 PCGs of Y. eos and other 18 species of Botiinae (Mao et al. Citation2014), a phylogenetic tree was constructed using neighbor-joining (NJ) method on Mega version 7.0 (US) with 1000 bootstrap replicates. The result showed that Botiinae was divided into two clusters (). Furthermore, the Yasuhikotakia and the Syncrossus clustered within the same clade. Y. eos has a closest relationship with Y. modesta, a species of the same genus.
tmdn_a_1842263_sm0294.tif
Download TIFF Image (1.2 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MT800510.
Additional information
Funding
References
- Frezal L, Leblois R. 2008. Four years of DNA barcoding: current advances and prospects. Infect. Genet. and Evol. 8(5):727–736.
- Kochzius M, Seidel C, Antoniou A, Botla SK, Campo D, Cariani A, Vazquez EG, Hauschild J, Hervet C, Hjörleifsdottir S, et al. 2010. Identifying fishes through DNA barcodes and microarrays. PLoS One. 5(9):e12620.
- Ma X, Huang F, Wang Z. 2014. The complete mitochondrial genome sequence of Acrossocheilus monticolus (Teleostei, Cypriniformes, Cyprinidae). Mitochondrial DNA. 25(4):245–246.
- Mao YT, Gan XN, Wang XZ. 2014. DNA barcodes and molecular phylogeny of Botiinae fishes based on the mitochondrial COI gene. Acta Hydrobiol Sin. 38(4):737–744.
- Nalbant TT. 2002. Sixty million years of evolution. Part one: family Botiidae (Pisces: Ostariophysi: Cobitidae). Trav Mus Hist Nat Grigore Antipa. 44:343–379.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Rainboth WJ. 1996. Fishes of the Cambodian Mekong. FAO species identification field guide for fishery purposes. Rome, Italy: FAO.
- Wang W, Li SK, Liu T, Wang X, Zeng S, Min WW, Zhou L. 2019. Characterization of the complete mitochondrial genome of an endangered fish Semilabeo obscurus (Cyprinidae; Labeoninae; Semilabeo). Conservation Genet Resour. 11(2):147–150.