Abstract
We determined the complete mitochondrial genomes of Crenidorsum turpiniae, a new record whitefly pest on tea-tree. The mitogenome of C. turpiniae is 15,427 bp long and consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes and a putative control region (GenBank: MN934936). The whole base composition of the heavy strand for A, C, G and T is 30%, 12.24%, 15.82% and 41.87%, respectively, with an AT bias (-16%). All PCGs use ATN as start codon (N, any nucleotide), except for NAD6 uses TTG. Most of the PCGs use TAA as a stop codon. The length of 16SrRNA and 12SrRNA gene are 1277 bp and 768 bp, respectively. Phylogenetic analysis indicated that C. turpiniae and Tetraleurodes acaciae had a closer genetic relationship.
Crenidorsum turpiniae, a new record whitefly pest of tea tree, which took place frequently in Meitan County of Guizhou Province in China, belongs to Aleyrodidae of Hemiptera. Nymphs prefer to feed on the underside of leaves using piercing-sucking mouthparts, which cause dark mildew in tea plants and seriously impact tea quality and yield. Adults like to inhabit together at the back of tender tea leaves, but can lay eggs at the whole plants (Meng et al. Citation2017).
Specimens of C. turpiniae were collected from a tea garden at Meitan County (27°44′21.89″N, 107°32′23.56″E) of Guizhou Province in China, in October 2018. We extracted genomic DNA from adults of C. turpiniae via the rapid extraction kit for genomic DNA from tissue cells (Aidlab Biotechnologies Co., Ltd). The puparium and adult specimens were deposited in Guizhou Tea Research Institute, Guizhou Academy of Agricultural Science, Guiyang, China, with the voucher specimen number of ET20 and EU21, for further study. The sequencing results were assembled and annotated using DNAstar, analyzed and adjusted them manually, and finally obtained the complete mitochondrial sequence – 37 typical invertebrate mitochondrial genes (13 protein-coding genes (PCGs), 22 tRNAs, and 2 rRNAs) and the A + T-rich region (D-loop). Annotated sequence of C. turpiniae mitogenome was submitted to GenBank with accession number MN934936.
The complete mitogenome of C. turpiniae is 15,427 bp. The nucleotide composition is AT-biased (−16%), with the respectively proportion as follow: A = 30% (4637), C = 12.24%(1889), G = 15.82%(2441) and T = 41.87%(6460). In the mt genome of C. turpiniae, a total of 30 bp overlaps have been found at 12 gene junctions and 12 small non-coding portions (intergenic spacers), ranging in size from 1 to 20 bp, and a large 1063 bp non-coding region (A + T-rich region) was identified also.
The PCGs, ranging in size from 148 bp(ATP8) to 1657 bp(ND5), with a whole length of 10,890 bp. All PCGs use ATN as start codon (N, any nucleotide), except for NAD6 use TTG. The COX1 gene use TAG as a stop codon, for the same time three PCGs (COX2, NAD5 and ATP8) use single T as a stop codon, all the rests use TAA as a stop codon. The tRNAs, ranging from 56 (Ser) to 70 (Lys and Glu) bp, similar to other aleyrodids (Chen et al. Citation2016; Wang et al. Citation2016). All tRNA genes have the conserved triple nucleotides recognizing corresponding codon. The length of 16SrRNA and 12SrRNA gene are 1277 bp and 768 bp, respectively.
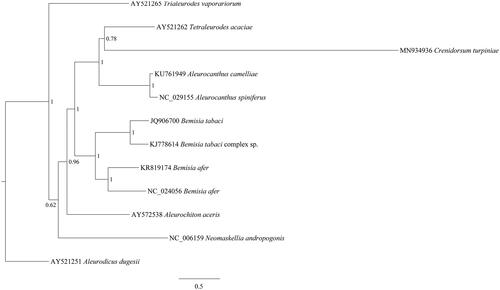
For understanding the evolutionary relationship of C. turpiniae in Aleyrodidae, the phylogenetic tree was constructed using MrBayes 3.2.0 (Ronquist et al. Citation2012) based on the nucleotide sequence of 13 PCGs of C. turpiniae and other 11 Aleyrodinae species (). We found that C. turpiniae and Tetraleurodes acaciae were clustered into one clade, and closely with Aleurocanthus camelliae and Aleurocanthus siniferus in relationship. The molecular data of C. turpiniae in this study provided important data in evolutionary analysis of Aleyrodidae phylogeny.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The sequence data generated in this study were openly available in GenBank (https://www.ncbi.nlm.nih.gov/nuccore/MN934936).
Additional information
Funding
References
- Chen SC, Wang XQ, Li PW, Hu X, Wang JJ, Peng P. 2016. The complete mitochondrial genome of Aleurocanthus camelliae: insights into gene arrangement and genome organization within the family aleyrodidae. IJMS. 17(11):1843.
- Meng ZH, Wang JR, Zhou XG, Li S, Yang W, Wang H, Zhou YF. 2017. A new insect pest Crenidorsum turpiniae (Takahashi, 1932) in tea plant (Camellia sinensis) and its biology. J Tea Sci. 37(6):638–644.
- Ronquist K, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Wang H-L, Xiao N, Yang J, Wang X-W, Colvin J, Liu S-S. 2016. The complete mitochondrial genome of Bemisia afer (hemiptera: Aleyrodidae). Mitochondrial DNA A. 27(1):98–99.