Abstract
Here, combining PacBio and Illumina sequencing data, we reported the complete chloroplast genome of the first Wuyi tea (Bohea), Camellia sinensis cv. Dahongpao (DHP) with very high economic value. The chloroplast genome was 157,077 bp in length, with a large single copy (LSC) region of 86,633 bp, a small single-copy (SSC) region of 18,282 bp, separated by two inverted repeat (IR) regions of 26,081 bp each. It contained a total of 137 genes, with an overall GC content of 37.29%. The phylogenetic analysis showed that DHP was sister to C. sinensis cv. Longjing.
Wuyi tea has a long history, formerly known as the trade name ‘Bohea’ in English. Because it was the first tea exported to the West, it was also called ‘Chinese tea’ in Europe in the seventeenth century. Wuyi tea is grown in rocky soil of Mountain Wuyi in northern Fujian, China, which is a UNESCO World Heritage site and considered the birthplace of oolong tea (Xiao Citation2017). As the most famous variety of Wuyi tea, DHP, collected from the original bushes of its variety, is among the most expensive teas in the world, and more valuable by weight than gold (Rose Citation2010).
Chloroplast (cp) genome, which is highly conserved in sequence and structure due to the non-recombinant, haploid, and uniparentally inherited nature, may provide useful information to further species identification and evolutionary study (Zeng et al. Citation2017). For further germplasm conservation, the complete cp genome sequence of DHP was sequenced using a combination of PacBio and Illumina sequencing platforms. The Illumina data was used to evaluate the complexity of the genome and correct the PacBio long reads. Total DNA was isolated from fresh leaves of an individual of DHP, collected from Mountain Wuyi (27°43′42.46″N, 118°0′14.40″E). All the voucher specimens of DHP were preserved in the Wuyi University herbarium, and DNA samples were stored at −80 °C at the Key Laboratory of Tea germplasm Genetic Resources of Wuyi University. PacBio and Illumina reads were mapped against the published cp genomes of Camellia sinensis var. sinensis (Accession number: KJ806281) (Huang et al. Citation2014) using CLC Genomics Workbench 11.0.1 software to filter out the cp reads and be de-novo assembled by guidance-based assembly approach. To annotate the cp genome, we used the initial annotation of cpGAVAS (Liu et al. Citation2012) and sequenced coordinates of annotated chloroplast genes available on NCBI using BLAST search (Acland et al. Citation2014). Annotation errors were corrected manually. Raw reads were deposited in the NCBI Sequence Read Archive (SRA: SRX8944642 and SRX8944643) and the final annotated cp genome sequence was deposited to NCBI GenBank (Accession number: MT773374).
The complete cp genome of DHP displayed the typical quadripartite structure of most angiosperm cp genomes, including the large single-copy (LSC, 86,633 bp), the small single-copy (SSC, 18,282 bp) and a pair of inverted repeats (IRa and IRb, 26,081 bp). The GC contents of the LSC, SSC, and IR regions individually, and of the cp genome as a whole, are 35.31, 30.53, 42.95 and 37.29%, respectively. It encoded a total of 113 unique genes, of which 22 were duplicated in the IR regions (ycf15 has four duplicates). Out of the 113 genes, there were 81 protein-coding genes, 28 tRNA genes, and 4 rRNA genes. 18 genes contained introns, 16 (twelve protein-coding and four tRNA genes) of which contained one intron and two of which (ycf3 and clpP) contained two introns.
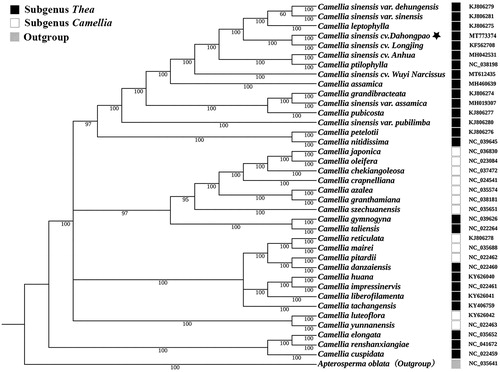
A phylogenetic analysis was performed based on complete cp genomes from 37 Camellia species representing two subgenera with Apterosperma oblata as the outgroup species. All of these 38 complete cp genomes were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). Bayesian inference analysis was carried out in MrBayes v.3.2.2 (Ronquist et al. Citation2012) based on the GTR substitution model, which was selected by the Akaike Information Criterion (Posada and Buckley Citation2004) as implemented in the program Modeltest v.3.7 (Posada and Crandall Citation1998). The result strongly supported that DHP was closely related to C. sinensis cv. Longjing (). In addition, our results also showed that not all subgenus Thea or subgenus Camellia were individually clustered according to Ming’s classification (Ming Citation2000), reflecting highly intricate phylogeny and taxonomy of Camellia (Lu et al. Citation2012).
Figure 1. Bayesian inference analysis based on 38 complete chloroplast genomes with Apterosperma oblata as an outgroup. The bootstrap support values (>50%) were shown above the branches. The position of Camellia sinensis cv. Dahonpao was marked with an asterisk and GenBank accession numbers were listed behind each species name. Species of subgenus Thea and subgenus Camellia were mark with the black box and the white box, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the US National Center for Biotechnology Information (NCBI database) at https://www.ncbi.nlm.nih.gov/, reference number: MT773374.
Additional information
Funding
References
- Acland A, Agarwala R, Barrett T, et al. 2014. Database resources of the National Center for Biotechnology Information Nucleic Acids Res. Nucleic Acids Res. 42:D7–D17.
- Huang H, Shi C, Liu Y, Mao S-Y, Gao L-Z. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14(1):151.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Lu H, Jiang W, Ghiassi M, Lee S, Nitin M. 2012. Classification of Camellia (Theaceae) species using leaf architecture variations and pattern recognition techniques. PLOS One. 7(1):e29704
- Ming TL. 2000. Monograph of the genus Camellia. Kunming: Yunnan Science and Technology Press.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 53(5):793–808.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Rose S. 2010. For all the tea in China: how England stole the world's favorite drink and changed history. Publisher: Viking Adult. Non-fiction, 272 pages. ISBN: 9780670021529. .
- Xiao KB. 2017. The taste of tea: material, embodied knowledge and environmental history in northern Fujian. China J Mater Cult. 22(1):3–18.
- Zeng S, Zhou T, Han K, Yang Y, Zhao J, Liu ZL. 2017. The complete chloroplast genome sequences of six Rehmannia species. Genes. 8(3):103.
