Abstract
We have determined the mitochondrial genome of A. gossypii isolated from Leonurus japonicus in Korea. The circular mitogenome of A. gossypii is 16,044 bp, including 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNAs, and a single control region of 797 bp. AT ratio is 83.8%. 88 SNPs and 4 INDELs (175 bp) were identified against mitogenome of A. gossypii isolated from cotton species. Especially, largest INDEL (170 bp) was in the control region. Phylogenetic trees show that four A. gossypii mitogenomes were clustered in one clade.
Aphis gossypii Glover, 1877 is one of the typical aphids showing wide range of hosts, especially for agriculture and horticultural species (Ebert and Cartwright Citation1997). More than 200 species have been identified as its host plants (Spradbery and Dvorak Citation2013). Usually, their host specificity is tightly linked to its genetic background; however, 7-day artificial feeding can alter their host specificity from cotton to cucumber (Ma et al. Citation2019), indicating that the genetic background of A. gossypii isolated from different host species is valuable to understand characteristics of A. gossypii. Till now three mitogenomes of A. gossypii isolated from cotton (Zhang S et al. Citation2016), Hibiscus syriacus (Park, Jung, et al. Citation2019), and Plantago asiatica (Bae et al. Citation2020) have been sequenced. Here, we presented the complete mitogenome of A. gosspii isolated from Leonurus japonicus in Korea.
Similar to the previous studies in which complete genomes were rescued from the sample containing multiple organisms (Bae et al. Citation2020; Park, Xi, Park, Lee Citation2020; Park, Xi, Park, Nam, et al. Citation2020; Oh et al. Citationin preparation), we sequenced and prepared the genomes from the L. japonicus sample with A. gossypii DNA (37.690627 N, 128.594893E; InfoBoss Cyber Herbarium (IN); INH-00026) extracted using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Sequencing library was constructed using Illumina TruSeq Nano DNA Library Preparation Kit (Illumina, San Diego, CA) following the manufacturer’s recommendations with around 350-bp DNA fragments. 4.19 Gbp raw sequences obtained from Illumina NovaSeq6000 (Macrogen Inc., South Korea) were filtered by Trimmomatic v0.33 (Bolger et al. Citation2014), de novo assembled by Velvet v1.2.10 (Zerbino and Birney Citation2008). Gaps were closed with SOAPGapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17, and SAMtools v1.9 (Li et al. Citation2009; Li Citation2013) under the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr/). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate mitogenome based on A. gossypii mitogenome (MN102349; Bae et al. Citation2020).
Aphis gossypii mitogenome (GenBank accession is MW013764) is 16,044 bp long, which is the second longest mitogenome among four available A. gossypii mitogenomes (Zhang S et al. Citation2016; Park Jung, et al. Citation2019; Bae et al. Citation2020). It contains 13 protein-coding genes, two rRNAs, and 22 tRNAs. Its nucleotide composition is AT-biased (A + T is 83.8%). Control region of 797 bp, which is longer than two A. gossypii mitogenomes (NC_024581 and MT430940), is found.
Eighty-eight single nucleotide polymorphisms (SNPs) and four insertions and deletions (INDELs) of which total length is 175 bp are identified against the mitogenome of A. gossypii isolated in cotton (NC_024581). Largest INDEL of which length is 170 bp is located in the control region, which is the most variable region in mitogenomes (Lee J et al. Citation2020; Zhang and Hewitt Citation1997). In addition, 35 SNPs and 4 INDELs (174 bp in total), 40 SNPs and 3 INDELs (3 bp) are identified against those isolated in H. syriacus (MN102349) and P. asiatica (MT430940), respectively. It is interesting that mitogenomes of A. gossypii isolated in L. japonicus and P. asiatica display the second lowest SNPs and significantly lowest total length of INDELs, indicating that control regions of both mitogenomes are similar to each other in comparison to the rest two mitogenomes.
Numbers of intraspecific variations identified against three previously sequenced mitogenomes are similar to those of Nilaparvata lugens (Choi et al. Citation2019; Park, Kwon, et al. Citation2019; Choi et al. Citation2020), Laodelphax striatellus (Park, Jung, et al. Citation2019; Seo, Jung, et al. Citation2019), and Spodoptera frugiperda (Seo, Lee, et al. Citation2019). However, they are smaller than those of Chilo suppressalis (Park, Xi, Kwon, et al. Citation2019) and are larger than those of Alphitobius diaperinus (Hong K-J et al. Citation2020) and Hipparchia autonoe (Lee Y-D et al. Citation2020).
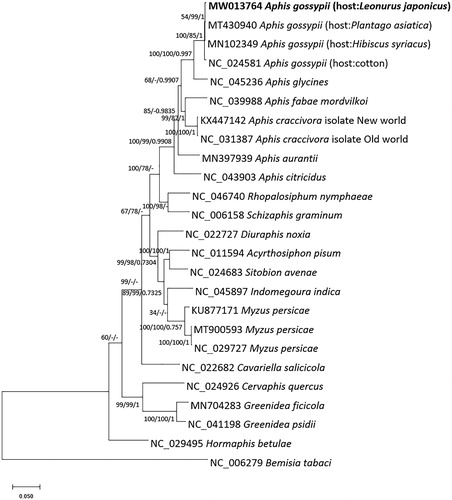
We inferred the phylogenetic relationship of 25 Aphididae mitogenomes, including four A. gossypii mitogenomes with one outgroup species, Bemisia tabaci (Tay et al. Citation2016). Multiple sequence alignment was conducted by MAFFT v7.450 (Katoh and Standley Citation2013). Bootstrapped maximum-likelihood, neighbor-joining, and Bayesian Inference trees were constructed using MEGA X (Kumar et al. Citation2018) and MrBayes v3.2.7a (Ronquist et al. Citation2012) based on multiple alignment of 25 mitogenomes. The result shows that our mitogenome was clustered with the other three mitogenomes of A. gossypii well (). However, there are many incongruencies among three different phylogenetic trees (), which is similar to previous analyses (Park, Xi, Kim, et al. Citation2019; Bae et al. Citation2020). In addition, the clade containing four A. gossypii mitogenomes displays low supportive values of maximum-likelihood and neighbor-joining trees (), requiring more phylogenetic analysis of A. gossypii. Our A. gossypii mitogenome can be used for understanding the potential relationship between host-specificity and mitogenomes together, additional mitogenomes originated from different hosts.
Figure 1. Maximum-likelihood (1000 bootstrap repeats), neighbor-joining (10,000 bootstrap repeats), and Bayesian Inference (1,100,000 generations) phylogenetic trees of 23 mitochondrial genomes of Aphididae and one outgroup: four Aphis gossypii (MW013764 in this study, MT430940 (Bae et al. Citation2020), MN102349 (Park J et al. Citation2019), and NC_024581 (Zhang S et al. Citation2016)), Aphis glycines (NC_045236; Song et al. Citation2019), Aphis fabae mordvilkoi (NC_039988; Voronova et al. Citation2020), Aphis caccivora (NC_031387 and KX447142; Sun et al. Citation2017), Aphis aurantia (MN397939; Wang Y et al. Citation2019), Aphis citricidus (NC_043903; Wei et al. Citation2019), Schizaphis graminum (NC_006158; Thao et al. Citation2004), Rhopalosiphum nymphaeae (NC_046740; Park, Kim, et al. Citation2020), Diuraphis noxia (NC_022727; De Jager et al. Citation2014), Myzus persicae (NC_029727, KU877171, and MT900593; Voronova et al. Citation2020; Cho et al., under revision), Indomegoura indica (NC_045897; Hong B et al. Citation2019), Sitobion avenae (NC_024683; Zhang B et al. Citation2016), Acyrthosiphon pisum (NC_011594), Cavariella salicicola (NC_022682; Wang Y et al. Citation2013), Cervaphis quercus (NC_024926; Wang Y et al. Citation2014), Greenidea psidii (NC_041198; Chen et al. Citation2019), Greenidea ficicola (MN704283; Liu et al. Citation2020), Hormaphis betulae (NC_029495; Li Y-Q et al. Citation2017), and Bemisia tabaci (NC_006279; Thao et al. Citation2004) as outgroup species. Phylogenetic tree was drawn based on the maximum-likelihood tree. The numbers above branches indicate bootstrap support values of maximum-likelihood, neighbor-joining, and Bayesian Inference phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MW013764 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA668988, SAMN16428189, and SRR12817477, respectively.
Additional information
Funding
References
- Bae Y, Park J, Lee W. 2020. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) isolated from Plantago asiatica in Korea. Mitochondrial DNA Part B. 5(3):2896–2898.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chen J, Wang Y, Qin M, Jiang L-Y, Qiao G-X. 2019. The mitochondrial genome of Greenidea psidii van der Goot (Hemiptera: Aphididae: Greenideinae) and comparisons with other Aphididae aphids. Int J Biol Macromol. 122:824–832.
- Choi NJ, Lee B-C, Park J, Park J. 2019. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in China (Hemiptera: Delphacidae): investigation of intraspecies variations between countries. Mitochondrial DNA Part B. 4(1):1677–1678.
- Choi NJ, Lee B-C, Park J, Park J. 2020. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in Guangxi province, China (Hemiptera: Delphacidae): identification of the origin of N. lugens migrated to Korea. Mitochondrial DNA Part B. 5(2):1960–1961.
- De Jager L, Burger N, Botha A. 2014. Complete mitochondrial genome of Diuraphis noxia (Hemiptera: Aphididae) from nine populations, SNP variation between populations, and comparison with other Aphididae species. African Entomol. 22(4):847–862.
- Ebert T, Cartwright B. 1997. Biology and ecology of Aphis gossypii Glover (Homoptera: aphididae). Southwestern Entomol. 22(1):116–153.
- Hong B, Zhang F, Hu Z-Q, Zhao H-Y. 2019. The complete mitochondrial genome of Indomegoura indica (Hemiptera: Aphididae). Mitochondrial DNA Part B. 4(1):882–883.
- Hong K-J, Ki W, Lee H, Park J, Lee W. 2020. The second complete mitochondrial genome of Alphitobius diaperinus Panzer, 1797 (Coleoptera: Tenebrionidae): investigation of intraspecific variations on mitochondrial genome. Mitochondrial DNA Part B. 5(3):2997–2999.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee J, Park J, Xi H, Park J. 2020. Comprehensive analyses of the complete mitochondrial genome of Figulus binodulus (Coleoptera: Lucanidae). J Insect Sci. 20(5):10.
- Lee Y-D, Lee J, Kim D-S, Park J, Xi H, Roh J, Kim D-S, Nam SJ, Kim S-K, Song J-Y, et al. 2020. The complete mitochondrial genome of Hipparchia autonoe (Esper, 1783)(Lepidoptera: Nymphalidae): investigation of intraspecific variations on mitochondrial genome. Mitochondrial DNA Part B. 5(2):1542–1544.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Li Y-Q, Chen J, Qiao G-X. 2017. Complete mitochondrial genome of the aphid Hormaphis betulae (Mordvilko) (Hemiptera: Aphididae: Hormaphidinae)). Mitochondrial DNA Part A. 28(2):265–266.
- Liu Q, Zhang H, Deng J, Lin X, Huang X. 2020. The complete mitochondrial genome of Greenidea ficicola (Hemiptera: Aphididae: Greenideinae), a pest of Ficus. Mitochondrial DNA Part B. 5(1):254–256.
- Ma L, Li M-Y, Chang C-Y, Chen F-F, Hu Y, Liu X-D. 2019. The host range of Aphis gossypii is dependent on aphid genetic background and feeding experience. PeerJ. 7:e7774.
- Oh S-H and Park J, The complete chloroplast genome of Leonurus japonicus Houtt, 1778 (Lamiaceae) isolated in Korea, in preparation.
- Park J, Jung JK, Ho Koh Y, Park J, Seo BY. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826; Hemiptera: Delphacidae) collected in a mid-Western part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2229–2230.
- Park J, Kim Y, Xi H, Park J, Lee W. 2020. The complete mitochondrial genome of Rhopalosiphum nymphaeae (Linnaeus, 1761) (Hemiptera: Aphididae). Mitochondrial DNA Part B. 5(2):1613–1615.
- Park J, Kwon W, Park J, Kim H-J, Lee B-C, Kim Y, Choi NJ. 2019. The complete mitochondrial genome of Nilaparvata lugens (stål, 1854) captured in Korea (Hemiptera: Delphacidae). Mitochondrial DNA Part B. 4(1):1674–1676.
- Park J, Xi H, Kim Y, Park J, Lee W. 2019. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) collected in Korean peninsula. Mitochondrial DNA Part B. 4(2):3007–3009.
- Park J, Xi H, Kwon W, Park C-G, Lee W. 2019. The complete mitochondrial genome sequence of Korean Chilo suppressalis (Walker, 1863)(Lepidoptera: Crambidae). Mitochondrial DNA Part B. 4(1):850–851.
- Park J, Xi H, Park J, Lee W. 2020. The complete mitochondrial genome of fungal endosymbiont, Ophiocordycipitaceae sp., isolated from Ricania speculum (Hemiptera: Ricaniidae). Mitochondrial DNA Part B. 5(2):1888–1889.
- Park J, Xi H, Park J, Nam SJ, Lee Y-D. 2020. Complete genome sequence of the Blochmannia endosymbiont of Camponotus nipponensis. Microbiol Resour Announc. 9(29), e00703–20.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Seo BY, Jung JK, Ho Koh Y, Park J. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826; Hemiptera: Delphacidae) collected in a southern part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2242–2243.
- Seo BY, Lee G-S, Park J, Xi H, Lee H, Lee J, Park J, Lee W. 2019. The complete mitochondrial genome of the fall armyworm, Spodoptera frugiperda Smith, 1797 (Lepidoptera; Noctuidae), firstly collected in Korea. Mitochondrial DNA Part B. 4(2):3918–3920.
- Song H, Donthu RK, Hall R, Hon L, Weber E, Badger JH, Giordano R. 2019. Description of soybean aphid (Aphis glycines Matsumura) mitochondrial genome and comparative mitogenomics of Aphididae (Hemiptera: Sternorrhyncha). Insect Biochem Mol Biol. 113:103208.
- Spradbery P, Dvorak L. 2013. Invasive species compendium. CABI. https://www.cabi.org/ISC
- Sun W, Huynh B-L, Ojo JA, Coates BS, Kusi F, Roberts PA, Pittendrigh BR. 2017. Comparison of complete mitochondrial DNA sequences between old and new world strains of the cowpea aphid, Aphis craccivora (Hemiptera: Aphididae). Agri Gene. 4:23–29.
- Tay W, Elfekih S, Court L, Gordon K, De Barro P. 2016. Complete mitochondrial DNA genome of Bemisia tabaci cryptic pest species complex Asia I (Hemiptera: Aleyrodidae). Mitochondrial DNA Part A. 27(2):972–973.
- Thao ML, Baumann L, Baumann P. 2004. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol Biol. 4(1):25.
- Voronova NV, Levykina S, Warner D, Shulinski R, Bandarenka Y, Zhorov D. 2020. Characteristic and variability of five complete aphid mitochondrial genomes: Aphis fabae mordvilkoi, Aphis craccivora, Myzus persicae, Therioaphis tenera and Appendiseta robiniae (Hemiptera; Sternorrhyncha; Aphididae). Int J Biol Macromol. 149:187–206.
- Wang Y, Ding M, Du Y, Huang A. 2019. Phylogenetic relationship and characterization of the complete mitochondrial genome of the black citrus aphid, Aphis aurantii (Hemiptera: Aphididae). Mitochondrial DNA Part B. 4(2):3567–3568.
- Wang Y, Huang X-L, Qiao G-X. 2013. Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLOS One. 8(10):e77511.
- Wang Y, Huang XL, Qiao GX. 2014. The complete mitochondrial genome of Cervaphis quercus (Insecta: Hemiptera: Aphididae: Greenideinae). Insect Sci. 21(3):278–290.
- Wei D-D, Lang N, Tao Y, He W, Tu Y-Q, Miao Z-Q, Yang L, Wang J-J. 2019. The mitochondrial genome of the brown citrus aphid Aphis (Toxoptera) citricidus: insights into the repeat regions in aphids and phylogenetic implications. Int J Biol Macromol. 136:531–539.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhang B, Zheng J, Liang L, Fuller S, Ma C-S. 2016. The complete mitochondrial genome of Sitobion avenae (Hemiptera: Aphididae). Mitochondrial DNA Part A. 27(2):945–946.
- Zhang D-X, Hewitt GM. 1997. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 25(2):99–120.
- Zhang S, Luo J, Wang C, Lv L, Li C, Jiang W, Cui J, Rajput LB. 2016. Complete mitochondrial genome of Aphis gossypii Glover (Hemiptera: Aphididae). Mitochondrial DNA Part A. 27(2):854–855.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
