Abstract
Leptopilina syphax (Hymenoptera: Figitidae) is a newly recorded species of parasitic wasp, and it attacks the larval stage of Drosophilidae, mainly the Drosophila species. Few works have been done in the basic study of L. syphax, including the data of mitochondrial genome. In this study, the complete mitochondrial genome of L. syphax (GeneBank accession number: MT649407) was sequenced using Illumina HiSeq X Ten system. The mitochondrial genome is 15,882bp long and comprises 13 protein-coding genes, 2 ribosomal RNA genes and 22 transfer RNA genes. Meanwhile, 26 genes are in majority strand, and the remaining 11 genes are in minority strand. The overall base composition is 41.7% for A, 6.0% for G, 13.6% for C, and 38.7% for T, with an A + T content of 80.4%, respectively. We also performed a phylogenetic analysis with other known mitochondrial genomes of some parasitic wasps. The results show that L. syphax is closely related to L. boulardi, which is another Drosophila parasitoid.
Complete mitochondrial genomes are useful tools for molecular evolution analysis and they have been wildly used to study phylogenetic relationship in both vertebrates and invertebrates (Ingman et al. Citation2000; Broughton et al. Citation2001; Mao et al. Citation2015). Mitochondrial genomes are treated as molecular markers for phylogenetic studies with many beneficial characteristics, such as lack of recombination, comparatively high mutation rate and the strictly maternal inheritance (Boore Citation1999). Leptopilina syphax is a newly recorded species of parasitic wasp and it attacks the larval stage of Drosophilidae, mainly the Drosophila species. Meanwhile, little has been done for its phylogenetic and evolution analysis.
Leptopilina syphax was collected by a net-trap method on May 2018 at Taizhou (28°50′N, 120°34′E), Zhejiang, China. The specimen (ZJUHJH_003) was stored in 100% ethanol and kept in the Parasitic Hymenoptera Collection of Institute of Insect Sciences, Zhejiang University. The DNA of L. syphax was isolated and purified using the standard phenol-chloroform method. The mitochondrial genome of L. syphax was sequenced by Illumina HiSeq X Ten system with the 150 paired-ends reading strategy. It was further annotated using the Geneious (version 11.0.4) and MITOS Web Server (Bernt et al. Citation2013).
The length of the complete mitochondrial genome of L syphax is 15,882 bp, and it contains 37 genes which includes 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNAgenes (rRNAs), and a putative control region (CR). Further analysis revealed that 26 genes are encoded on the majority strand, while the remaining 11 genes are encoded on the minority strand. The gene order in the L. syphax mitochondrial genome is very similar with another Leptopilina species, L. boulardi (Oliveira et al. Citation2016). The overall base composition is 41.7% for A, 6.0% for G, 13.6% for C, and 38.7% for T, with an A + T content of 80.4%. Three start codons for PCGs are used: ATA (nad2, nad1 and nad5); ATT (cox1, atp8, cox3, nad4l and cob); ATG (cox2, atp6, nad3, nad4 and nad6). 10 PCGs use a TAA stop codon and 3 PCGs (cox3, nad3 and cob) use a TAG stop codon. The 22 tRNAs genes vary from 58 bp to 73 bp in length, and the secondary structure of tRNAs are typical clover-leaf structures as with other insects. The rrnL was located next to rrnS, and they both were located between trnD and trnC. The length of rrnL and rrnS was 1316 bp and 817 bp, respectively.
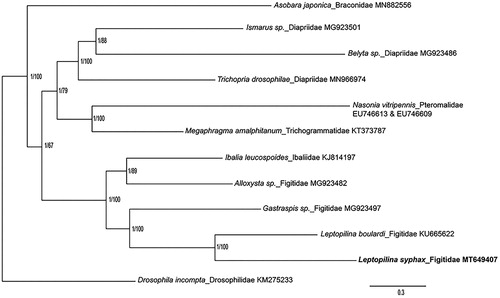
Till now, 50 hymenopterous parasitoid species have been founded to attack Drosophilid species worldwide, and the majority of which are in the following four families including Braconidae, Figitidae, Diapriidae and Pteromalidae. We performed the phylogenetic analysis of L. syphax with some other parasitoids from the four families and with Megaphragma amalphitanum (Trichogrammatidae) and Ibalia leucospoides (Ibaliidae) as well, which are closely related with Pteromalidae and Figitidae, respectively. (Oliveira et al. Citation2008, Citation2016; Li et al. Citation2016; Tang et al. Citation2019; Zhang, Li, et al. Citation2020; Zhang, Pan, et al. Citation2020). The sequences were aligned using MAFFT v7.271, and the phylogenetic tree was constructed by CIPRES (https://www.phylo.org/) using RAxML-HPC2 on XSEDE with bootstrap 1000 and MrBayes on XSEDE (). Phylogenetic analysis showed that L. syphax is closely related to L. boulardi, another Drosophila parasitoid in Leptopilina genus. This study would further clarify our understanding of the phylogenetic relationship of the Figitidae family.
Figure 1. Phylogenetic relationships among selected parasitoids inferred from nucleotides of 13 PCGs and 2 rRNAs using Bayesian and maximum-likelihood (ML) methods (GenBank accession numbers provided). The Bayesian posterior probabilities (PP) and bootstrap support (BS) are marked beside the nodes. Drosophila incompta was set as outgroup.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT649407.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69 (2):313–319.
- Boore J. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27 (8):1767–1780.
- Broughton R, Milam J, Roe B. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11(11):1958–1967.
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. 2000. Mitochondrial genome variation and the origin of modern humans. Nature. 408(6813):708–713.
- Li Q, Wei S, Tang P, Wu Q, Shi M, Sharkey MJ, Chen X. 2016. Multiple lines of evidence from mitochondrial genomes resolve phylogenetic relationships of parasitic wasps in Braconidae. Genome Biol Evol. 8(9):2651–2662.
- Mao M, Gibson T, Dowton M. 2015. Higher-level phylogeny of the Hymenoptera inferred from mitochondrial genomes. Mol Phylogenet Evol. 84:34–43.
- Oliveira D, Gomes T, Loreto E. 2016. The rearranged mitochondrial genome of Leptopilina boulardi (Hymenoptera: Figitidae), a parasitoid wasp of Drosophila. Genet Mol Biol. 39(4):611–615.
- Oliveira D, Raychoudhury R, Lavrov D, Werren J. 2008. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp nasonia (hymenoptera: pteromalidae). Mol Biol E. 25 (10):2167–2180.
- Tang P, Zhu J, Zheng B, Wei S, Sharkey M, Chen X, Vogler A. 2019. Mitochondrial phylogenomics of the Hymenoptera. Mol Phylogenet Evol. 131:8–18.
- Zhang X, Li C, Pan Z, Zhu J, Wang Z, Shi M, Chen X, Huang J. 2020. The complete mitochondrial genome of Asobara japonica (Hymenoptera: Braconidae. ). Mitochondrial DNA Part B. 5 (2):1279–1281.
- Zhang X, Pan Z, Chen J, Zhu J, Zhou S, Pang L, Shi M, Chen X, Huang J. 2020. The complete mitochondrial genome of Trichopria drosophilae (Hymenoptera: Diapriidae). Mitochondrial DNA Part B. 5(3):2391–2393.
