Abstract
Saussurea inversa Raab-Straube is a small perennial and polycarpic herb that is distributed on the rocky slopes of the Qinghai-Tibetan plateau (QTP) at an altitude of 3700–5400 meters. It has a chloroplast genome structure similar to that of other species of Saussurea. It is 152,102 bp in size, including a large single-copy (LSC) region of 83,450 bp, a small single-copy (SSC) region of 18,286 bp and a pair of inverted repeats (IRs) of 25,183 bp. The chloroplast genome of S. inversa encodes 113 genes, containing 80 protein-coding genes (PCGs), 29 tRNA and four rRNA genes. Phylogenetic analysis shows that S. inversa is clustered with S. pseudosimpsoniana and S. laniceps.
Saussurea inversa (subgenus Eriocoryne, Cynareae, Asteraceae) is a perennial and polycarpic herb, which grows in high-altitude alpine areas of QTP (Shi et al. Citation2011). It is mainly distributed in Tibet, Sichuan, Qinghai and Yunnan of China, which are thought to be the diversity center and origin place of genus Saussurea (Xu et al. Citation2019). Studies have shown that the differentiation of various species of Saussurea has a close relationship with the last dramatic uplift of the QTP (Liu et al. Citation2006; Wang et al. Citation2009). Here, we first sequenced and reported the complete chloroplast genome of S. inversa (GenBank accession number: MT554929).
In the present study, the fresh samples of S. inversa were collected from Qilian Mountain of Qinghai, China (98°44′56.3″, 38°46′40.6″). The voucher specimen of S. inversa was deposited in the Qinghai-Tibetan Plateau Museum of Biology, Northwest Institute of Plateau Biology, Chinese Academy of Sciences, with the collection number of QPMB 0334381. Modified CTAB method (Idress Hamad Citation2011) was used to extract the genomic DNA. The quantity and quality of the DNA were evaluated using the Nanodrop 2000 and 1% agarose gel electrophoresis, respectively. Total DNA was used to generate libraries with an average insert size of 350 bp, which were sequenced using Illumina HiSeq platform provided by Genepioneer Biotechnologies (Nanjing, China). Finally, about 4.05 GB of clean data were generated and the SPAdes (Bankevich et al. Citation2012) was used to genome assembly, based on the clean data. The Geseq online software (Tillich et al. Citation2017) was used to annotate the functional genes. The results showed that the complete chloroplast genome of S. inversa (152,102 bp, 37.73% GC content) possessed quadripartite structure, containing a LSC (83,450 bp, 35.83% GC content) region, an SSC (18,286 bp, 31.51% GC content) region and a pair of IRs (25,183 bp, 43.13% GC content) regions. Totally, 80 PCGs, 29 tRNA and four rRNA genes were identified, in which 12 genes (atpF, ndhA, ndhB, rpl2, rps16, rpoC1, trnA-UGC, trnG-UUC, trnI-UAA, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron, and two genes (clpP and ycf3) contained two introns.
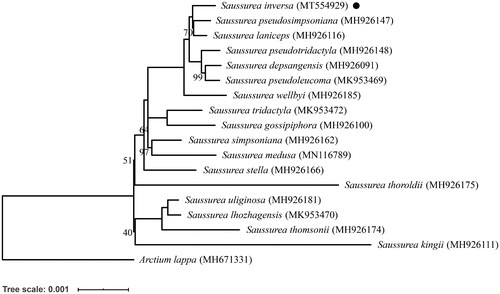
The Maximum Likelihood (ML) phylogenetic tree of 17 species of subgenus Eriocoryne and one outgroup species was constructed using IQ-tree version 1.6.12 (Trifinopoulos et al. Citation2016) under the best model of K3Pu + F+R3 and bootstrapped with 1000 replicates (). The tree was visualized using the iTOL online tool (Letunic and Bork Citation2016). According to the results, S. inversa was closely related to S. pseudosimpsoniana and S. laniceps.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The raw data of sequencing is openly available in Figshare at https://figshare.com/articles/SRR11862870/12470204/1.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Idress Hamad A. 2011. Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pak J Biol Sci. 21:998–999.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–245.
- Liu JQ, Wang YJ, Wang AL, Hideaki O, Abbott RJ. 2006. Radiation and diversification within the Ligularia–Cremanthodium–Parasenecio complex (Asteraceae) triggered by uplift of the Qinghai-Tibetan Plateau. Mol Phylogenet Evol. 38(1):31–49.
- Shi Z, Chen YL, Chen YS, Lin YR, Liu SW, Ge XJ, Gao TG, Zhu SX, Liu Y, Yang QE, et al. 2011. Asteraceae (Compositae) [family introduction, glossary, systematic list, and key to tribes]. In: Wu ZY, Raven, H. P, and Hong DY, eds. Flora of China (Asteraceae). Beijing: Science Press; St. Louis: Missouri Botanical Garden Press. Vol 20-21: 62.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–235.
- Wang YJ, Susanna A, Von Raab-Straube E, Milne R, Liu JQ. 2009. Island-like radiation of Saussurea (Asteraceae: Cardueae) triggered by uplifts of the Qinghai-Tibetan Plateau. Biol J Linnean Soc. 97(4):893–903.
- Xu L-S, Herrando-Moraira S, Susanna A, Galbany-Casals M, Chen Y-S. 2019. Phylogeny, origin and dispersal of Saussurea (Asteraceae) based on chloroplast genome data. Mol Phylogenet Evol. 141:106613.