Abstract
The complete mitogenome sequence of Sinogastromyzon szechuanensis was obtained using next generation sequencing and confirmed via overlap-PCR method. The genome was 16,565 bp in length and consisted of 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 23 transfer RNA genes and 1 control region. The overall nucleotide composition of heavy strand was 30.38% A (5033), 16.62% G (2753), 25.25% T (4182) and 27.75 (4597), with a slight A–T skew (55.63%), which is most obvious in the D-loop and most transfer RNA genes. Mitochondrial genome analyses based on ML analyses yielded identical phylogenetic trees.
Sinogastromyzon szechuanensis Fang (Ding Citation1994), belonging to Balitoridae of Cypriniform, is a small freshwater and endemic to the upper Yangtze River, China. Being different from other Cypriniform fish, S. szechuanensis is of flattened body and capable of jumping among the stones of waterfall or mountain streams (Jin-Ming et al. Citation2011; Wu et al. Citation2013). They are of high economic value for their good taste. Unfortunately, they were in danger for their high price and shrunken habitat. There were only a few documents or researches focus on them. It will be a tremendous priority to take action to protect and use them carefully.
The S. szechuanensis were collected from Neijiang, Sichuan, China (29°36′41.93″N, 105°01′47.67″E) and stored at Aquatic Science and Technology Institution Herbarium (Accession number: SC20180913HXQ02). Total genomic DNA of liver was extracted using Magnetic Animal Tissue Genomic DNA kit of Tiangen Biotech (Beijing) Co., Ltd (Yu et al. Citation2019). The genomic DNA (DNA label: S. szechuanensis 001) were stored in an ultralow refrigerator (–70 °C) of the Herbarium.
A genomic DNA library was established and sequenced to assembly the whole mitochondrial genome using next-generation sequencing method (Illumina HiSeq platform) (Asem et al. Citation2018). Quality check for sequencing data was done by FastQC and the fragments sequences were assembled and mapped using SPAdes (Bankevich et al. Citation2012). Thirty-five pairs of PCR primers were designed to amplify the whole mitogenome sequence based on the assembled DNA sequences to affirm the assembled results.
The complete mitogenome of S. szechuanensis is 16,565 bp in length (NCBI accession ID: MN241814) and of conserved structural organization, which consisted of 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 23 transfer RNA genes and 1 control region displacement loop (D-loop). The overall nucleotide composition of heavy strand was 30.38% A (5033), 16.62% G (2 753), 25.25% T (4 182) and 27.75 (4 597), with a slight A–T skew (55.63%), which is most obvious in the D-loop and most transfer RNA genes. Except for seven RNAs (tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu and tRNA-Pro) and one protein (ND6), which were encoded on the L-strand, most elements were encoded on the heavy strand (H-strand). 12 s rRNA (944 bp) and 16 s rRNA (1 508 bp) were separated by a gap of 151 bp, while tRNA-Val was encoded within the gap.
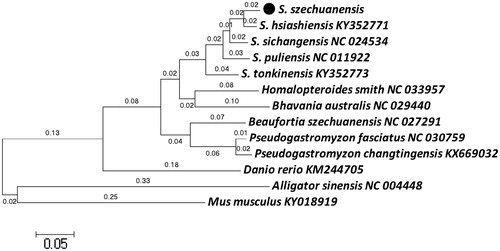
The maximum-likelihood (ML) phylogenetic (Zhu et al. Citation2018) tree was constructed with MEGA 6.0 program based on 13 complete mitochondrial genome sequences (Asem et al. Citation2018) (). The results showed a close relationship among S. szechuanensis, S. hsiashiensis (KY352771), S. sichangensis (NC_024534), S. puliensis (NC_011922), S. tonkinensis (KY352773) and a relative loose relationship between S. szechuanensis and Danio retio (KM244705). In conclusion, this complete mitochondrial genome would establish a solid foundation for future population geography and conservation genetic studies of S. szechuanensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MN241814.1
Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA667134, https://www.ncbi.nlm.nih.gov/sra/ SRR12767438, and SAMN16356296, respectively.
Additional information
Funding
References
- Asem A, Wang PZ, Li WD. 2018. The complete mitochondrial genome of the Asian river pipefish Doryichthys boaja (Actinopterygii; Syngnathiformes; Syngnathidae) obtained using next-generation sequencing. Mitochondrial DNA Part B. 3(2):776–777.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Ding RH. 1994. The fishes of Sichuan, China. Chengdu: SichuanPublishing House of Science and Technology.
- Jin-Ming WU, Wang QQ, Liu F, Liu CC, Wang JW. 2011. Early development of Sinogastromyzon szechuanensis in the Chishui River. Sichuan J Zool 527- 529, 536
- Wu J, Li L, Du H, Zhang H, Wang C, Wei Q. 2013. Length-weight relations of 14 endemic fish species from the upper Yangtze River Basin, China. Acta Icth Piscat. 43(2):163–165.
- Yu P, Zhou L, Zhou XY, Yang WT, Zhang J, Zhang XJ, Wang Y, Gui JF. 2019. Unusual AT-skew of Sinorhodeus microlepis mitogenome provides new insights into mitogenome features and phylogenetic implications of bitterling fishes. Int J Biol Macromol. 129:339–350.
- Zhu K, Gong L, Jiang L, Liu L, Lu Z, Liu B-J. 2018. Phylogenetic analysis of the complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae). Mitochondrial DNA Part B. 3(2):536–537.