Abstract
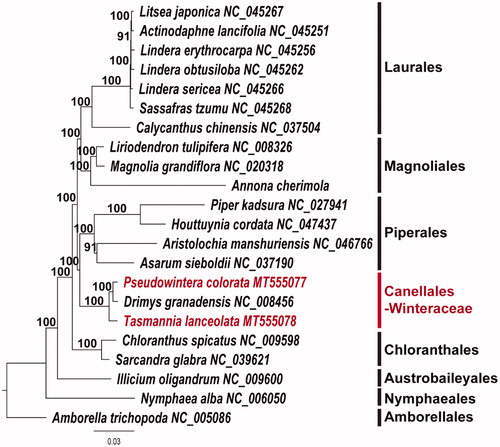
In this study, we report the complete plastome sequences of two Winteraceae taxa, Pseudowintera colorata (MT555077) and Tasmannia lanceolata (MT555078). Both plastomes show typical quadripartite structure. The plastome size of P. colorata is 161,675 bp, which consists of 89,583 bp large single-copy (LSC), 18,606 bp small single-copy (SSC), and 26,743 bp inverted repeat (IR) regions. The plastome size of T. lanceolata is 160,424 bp, which consist of 88,589 bp LSC, 18,351 bp SSC, and 26,742 bp IR regions. Both plastomes contain 113 genes, including 79 protein-coding, 30 tRNA, and four rRNA genes. Sixteen genes contain one intron and two genes (clpP and ycf3) have two introns. Ninety-three and 89 simple sequence repeat (SSR) loci are scattered in the P. colorata and T. lanceolata plastomes, respectively. Our phylogenetic tree shows the relationship of (T. lanceolate (P. colorata, Drimys granadensis)) in the Winteraceae. The Canellales (incl. Winteraceae) are the sister group of Piperales.
Pseudowintera colorata and Tasmannia lanceolata are native to New Zealand and Southeastern Australia, respectively. They belong to the family Winteraceae in the order Canellales (APG IV Citation2016). Winteraceae consists of two subfamilies, five genera, and approximately 105 species (Christenhusz and Byng 2016). Pseudowintera and Tasmannia belong to the subfamily Winteroideae and consist of approximately three and 50 species, respectively.
The leaves of P. colorata and T. lanceolata were ground into powder in liquid nitrogen and total DNAs were extracted using the G-spinTM IIp for Plant Genomic DNA Extraction Kit (iNtRON Biotechnology, Gyeonggi-do, Korea). Two voucher specimens were deposited in the Korea University Herbarium (KUS acc. nos. TN2020-0006 and TA2019-0017) and genomic DNAs are deposited in the Plant DNA Bank in Korea (PDBK acc. nos. 2020-0006 and 2019-0017). Two complete plastome sequences were generated using an Illumina MiSeq platform (Illumina Inc., San Diego, CA). De novo assemblies and annotations of plastomes were performed using the Geneious version 11.1.5 (Biomatters Ltd., Auckland, New Zealand; Kearse et al. Citation2012), National Center for Biotechnology Information (NCBI) BLAST, and tRNAscan-SE programs (Lowe and Eddy Citation1997). The average plastome coverage of P. colorata and T. lanceolata is 661× and 2006×, respectively. The simple sequence repeats (SSRs) were detected by the Phobos version 3.3.12 program (Leese et al. Citation2008) in the Geneious version 11.1.5. For the phylogenetic analysis, we selected and downloaded 20 related complete plastome sequences based on the APG IV system (APG IV Citation2016) from the NCBI database.
The plastome size of P. colorata is 161,675 bp, which consist of 89,583 bp large single-copy (LSC) region, 18,606 bp small single-copy region (SSC), and 26,743 bp inverted repeat (IR) region. The plastome size of T. lanceolata is 160,424 bp, which consists of 88,589 bp LSC region, 18,351 bp SSC region, and 26,742 bp IR region. They are similar to the Drimys granadensis (160,604 bp, NC008456) plastome reported in previous studies (Cai et al. Citation2006). They show a typical quadripartite structure. Both plastomes hold 113 unique genes, including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. Six protein-coding, seven tRNA, and four rRNA genes are duplicated in the IR regions. The average A-T contents of the two plastomes are 61.1% in P. colorata and 61.2% in T. lanceolata, respectively. Sixteen genes contain one intron and two genes, ycf3 and clpP, have two introns. A total of 93 SSR loci are distributed throughout the P. colorata plastome. Among these, 78, 10, and 5 are mono-SSR, di-SSR, and tri-SSR loci, respectively. A total of 89 SSR loci including 71 mono-, 14 di-, and 4 tri-SSRs are scattered in the T. lanceolata plastome.
To estimate the phylogenetic relationships of two species, we constructed a maximum likelihood (ML) tree using 22 basal angiosperm taxa. Phylogenetic analysis was performed on a data set that included 79 protein-coding genes and four rRNA genes from the 22 selected taxa using RAxML version 8.2.12 in CIPRES webserver (Stamatakis Citation2014) under GTR + G + I model with 1000 bootstrap replicates. The 83 gene sequences (79,005 bp in length) were aligned with the MUSCLE program using Geneious version 11.1.5 (Biomatters Ltd. Auckland, New Zealand; Kearse et al. Citation2012). The resulting tree supports the monophyly of three members of Winteraceae by 100% bootstrap value. The tree shows the (T. lanceolate (P. colorata, D.granadensis)) relationship within the Winteraceae. The Canellales (incl. Winteraceae) were the sister group of Piperales (). This relationship is also supported by the previous studies (Karol et al. Citation2000; Doust and Drinnan Citation2004). The complete plastome is rare in the basal lineages of angiosperm. Only one sequence in Canellales was available before our report. Therefore, the complete plastome sequences of P. colorata and T. lanceolata will provide a useful resource for the evolutionary studies of Canellales.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the finding of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT555077 for Pseudowintera colorata and MT555078 for Tasmannia lanceolata.
Additional information
Funding
References
- APG IV. 2016. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20.
- Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, dePamphilis CW, Boore JL, Jansen RK. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 6:77.
- Christenhusz MJ, Byng JW. 2016. The number of known plants species in the world and its annual increase. Phytotaxa. 261(3):201–217.
- Doust AN, Drinnan AN. 2004. Floral development and molecular phylogeny support the generic status of Tasmannia (Winteraceae). Am J Bot. 91(3):321–331.
- Karol KG, Suh Y, Schatz GE, Zimmer EA. 2000. Molecular evidence for the phylogenetic position of Takhtajania in the Winteraceae: inference from nuclear ribosomal and chloroplast gene spacer sequences. Ann Missouri Bot Gard. 87(3):414–432.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Leese F, Mayer C, Held C. 2008. Isolation of microsatellites from unknown genomes using known genomes as enrichment templates. Limnol Oceanogr Methods. 6(9):412–426.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.