Abstract
The genus Tanyptera Latreille, Citation1804 is recorded from Shandong Province, China for the first time with T. (T.) hebeiensis Yang et Yang, Citation1988 found in Mount Kunyu, Shandong. In this study, we report the complete mitochondrial genome sequence of T. (T.) hebeiensis, representing the first mitochondrial genome of the subfamily Ctenophorinae (Diptera: Tipulidae), which is a circular molecule of 15,888 bp with an AT content of 77.6%. The mitochondrial genome contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and a non-coding region. Gene overlaps are found at nine gene junctions, ranging from 1 to 8 bp in length. The canonical mitochondrial start codons for invertebrate mitochondrial genomes are found in 12 PCGs, except for COI which uses the uncanonical start codons TCG. Stop codons of 10 PCGs are invariably complete TAA and TAG, while COII, ND4, and ND5 end with a single thymine stop codon. Phylogenetic analysis reveals that the Pediciidae is a sister group to the remaining Tipuloidea, the Cylindrotomidae has a sister-group relationship with the Tipulidae, and the Limoniidae is not a monophyletic clade.
The genus Tanyptera Latreille, Citation1804 is a small group in crane-flies with 27 known species in the world, of which 22 species were known from the Palearctic Region, four species were known from the Oriental Region and only one species was known from the Nearctic Region (Oosterbroek Citation2020). There were 13 known species of the genus Tanyptera from China distributed in the following 13 provinces, municipalities or autonomous regions: Sichuan (3 species), Hubei (3), Fujian (2), Hebei (2), Gansu (2), Shaanxi (2), Zhejiang (1), Jiangxi (1), Heilongjiang (1), Anhui (1), Neimenggu (1), Beijing (1), Ningxia (1). In the present paper, the genus Tanyptera is recorded from Shandong Province, China for the first time with the species T. (T.) hebeiensis Yang et Yang, Citation1988 (specimens examined: 10 males 8 males, China, Shandong, Muping, Mount Kunyu, 2019.VII, Chuande Zhao, malaise trap) (). The mitochondrial genome of T. (T.) hebeiensis is also sequenced and analyzed.
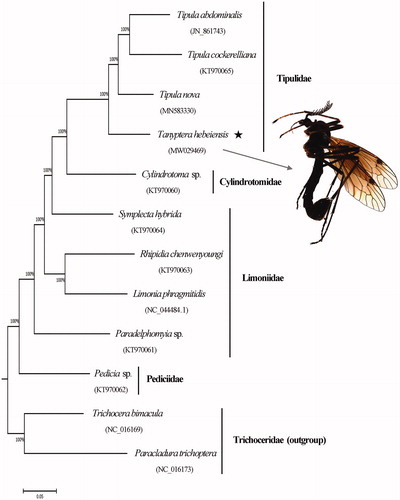
Figure 1. Phylogenetic tree of Tipuloidea based on whole mitochondrial genomes using maximum-likelihood analysis. Numbers above the branches are bootstrap percentages. GeneBank accession numbers of each species were listed in the tree.
So far a large number of mitochondrial genomes have been widely used for reconstructing phylogenetic relationships in many insect groups (Beckenbach Citation2012; Timmermans and Vogler Citation2012; Wang et al. Citation2012; Caravas and Friedrich Citation2013; Li et al. Citation2013; Li et al. Citation2015; Zhang et al. Citation2016; Kang et al. Citation2017; Wang et al. Citation2017; Zhang et al. Citation2019). There are about 700 complete or nearly complete lower dipteran mitochondrial genomes available in GenBank, of which four are from the family Tipulidae (Beckenbach Citation2012; Zhang et al. Citation2016; Zhao et al. Citation2019). However, they are all from the genus Tipula Linnaeus, Citation1758 of the subfamily Tipulinae (Tipulidae). Here, we report the first complete mitochondrial genome sequence of the subfamily Ctenophorinae (Tipulidae), T. (T.) hebeiensis mitochondrial genome, which will provide an insight into the phylogeny.
The specimen of T. (T.) hebeiensis used in this study was collected from Mount Kunyu in Shandong province, China (37°16′30″N, 121°45′57″E; 200 m) and stored in the Entomological Museum of Qingdao Agricultural University, China (No. TIP0003). The total DNA was extracted from the thoracic muscle using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China). The mixed DNA library was constructed with Illumina hiseq by Berrygenomics Co., Ltd (Beijing, China) and the fragment COI was amplified as a ‘bait’ sequence to identify the target mitochondrial genome (Gillett et al. Citation2014). The standard PCR reactions were sequenced by primers designed by Simon et al. (Citation2006). BLAST searches were conducted with BioEdit 7.0.5.3 for the bait sequence against mitochondrial genome assemblies. The sequence was annotated following the method proposed by Cameron (Citation2014). The transfer RNA (tRNA) genes were identified with tRNAscan-SE v2.0. The ribosomal RNA (rRNA) genes were detected by alignment with homologous sequences obtained from the published species. The boundaries of protein-coding genes (PCGs) were identified based on open reading frames provided by ORF Finder (https://www.ncbi.nlm.nih.gov/gorf/gorf.html). Maximum-likelihood analysis was conducted by MEGA7 (Kumar et al. Citation2016).
The total length of the mitochondrial genome of T. (T.) hebeiensis (GenBank accession no. MW029469) is 15,888 bp and the nucleotide composition of the mitochondrial genome is AT biased (A + T contents: 77.6%). It contains 13 PCGs, 22 tRNAs, 2 rRNAs and a 1065 bp long non-coding region. The gene order shows a conserved arrangement pattern: 23 genes are oriented on the majority strand and 14 genes are transcribed on the minority strand. In the T. (T.) hebeiensis mitochondrial genome, gene overlaps are found at nine gene junctions, ranging from 1 to 8 bp in length. The longest overlaps exist between tRNATrp and tRNACys. Twelve small non-coding intergenic spacers are found in the T. (T.) hebeiensis mitochondrial genome, ranging from 2 to 27 bp in length. The largest non-coding intergenic spacers is between tRNAGlu and tRNAPhe. The canonical mitochondrial start codons (ATN) for invertebrate mitochondrial genomes (Wolstenholme Citation1992) are found in 12 PCGs of the T. (T.) hebeiensis mitochondrial genome, except for COI which uses the uncanonical start codons TCG. Stop codons of 10 PCGs in T. (T.) hebeiensis mitochondrial genome are invariably complete TAA and TAG, while COII, ND4, and ND5 end with a single thymine stop codon. Twenty-two typical tRNAs in the arthropod mitochondrial genomes are found in the T. (T.) hebeiensis mitochondrial genome, ranging from 62 to 72 bp. The length of the rRNAs in T. (T.) hebeiensis mitochondrial genome are determined to be 1323 bp for lrRNA and 791 bp for srRNA.
The phylogenetic tree () in this study indicates that the family Pediciidae is sister-group to the remaining Tipuloidea. The sister-group relationship between the families Cylindrotomidae and Tipulidae is strongly supported, which is concordant with Starý (Citation1992), Ribeiro (Citation2008), Petersen et al. (Citation2010), Zhang et al. (Citation2016) and Kang et al. (Citation2017). The family Limoniidae is not supported as a monophyletic clade in this study, which is also accepted by Ribeiro (Citation2008), Petersen et al. (Citation2010) and Zhang et al. (Citation2016).
Acknowledgments
The authors express our sincere thanks to Ding Yang (Beijing) and Fan Song (Beijing) for great help during the study. The authors are also very grateful to Yan Li (Shenyang) for identifying the specimens.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MW029469. Associated BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA669920, https://www.ncbi.nlm.nih.gov/sra/SRR12880699, and SAMN16481077, respectively. Other GeneBank accession numbers were listed as follows: Cylindrotoma sp. (KT970060), Symplecta (Symplecta) hybrida (KT970064), Limonia phragmitidis (NC_044484.1), Paracladura trichoptera (NC_016173), Paradelphomyia sp. (KT970061), Pedicia sp. (KT970062), Rhipidia (Rhipidia) chenwenyoungi (KT970063), Tipula (Nippotipula) abdominalis (JN_861743), Tipula (Acutipula) cockerelliana (KT970065), Tipula (Yamatotipula) nova (MN583330), and Trichocera bimacula (NC_016169).
Additional information
Funding
References
- Beckenbach AT. 2012. Mitochondrial genome sequences of Nematocera (Lower Diptera): evidence of rearrangement following a complete genome duplication in a Winter Crane fly. Genome Biol Evol. 4(2):89–101.
- Cameron SL. 2014. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst Entomol. 39(3):400–411.
- Caravas J, Friedrich M. 2013. Shaking the Diptera tree of life: performance analysis of nuclear and mitochondrial sequence data partitions. Syst Entomol. 38(1):93–103.
- Gillett C, Alex CP, Timmermans M, Jordal BH, Emerson BC, Vogler AP. 2014. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol Biol Evol. 31(8):2223–2237.
- Kang Z, Zhang X, Ding S, Tang C, Wang Y, de Jong H, Cameron SL, Wang M, Yang D. 2017. Transcriptomes of three species of Tipuloidea (Diptera, Tipulomorpha) and implications for phylogeny of Tipulomorpha. PLOS One. 12(3):e0173207.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Latreille PA. 1804. Tableau methodique des Insectes. In: Societe de Naturalistes et d Agriculteurs: Nouveau dictionnaire d histoire naturelle, appliquee aux arts, principalement a l agriculture et a l economie rurale et domestique. Tableaux methodiques d histoire naturelle. Paris; Vol. 24(3); p. 1–238.
- Li H, Shao R, Song F, Zhou X, Yang Q, Li Z, Cai W. 2013. Mitochondrial genomes of two Barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): contrasting rates in mitochondrial gene rearrangement between major lineages of Psocodea. PLOS One. 8(4):e61685.
- Li H, Shao R, Song N, Song F, Jiang P, Li Z, Cai W. 2015. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci Rep. 5:8527.
- Linnaeus C. 1758. Systema naturae per regna tria naturae, secundum classes, ordines, genera, species, cum caracteribus, differentiis, synonymis, locis. Ed. 10. Laurentii Salvii, Holmiae [= Stockholm]. 1: i–iv, 1–824.
- Oosterbroek P. 2020. Catalogue of the Craneflies of the World (Diptera, Tipuloidea: Pediciidae, Limoniidae, Cylindrotomidae, Tipulidae); [accessed 2020 Sep 20]. https://ccw.naturalis.nl/.
- Petersen MJ, Bertone MA, Wiegmann BM, Courtney GW. 2010. Phylogenetic synthesis of morphological and molecular data reveals new insights into the higher-level classification of Tipuloidea (Diptera). Syst Entomol. 35(3):526–545.
- Ribeiro GC. 2008. Phylogeny of the Limnophilinae (Limoniidae) and early evolution of the Tipulomorpha (Diptera). Invert Syst. 22(6):627–694.
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 37(1):545–579.
- Starý J. 1992. Phylogeny and classification of Tipulomorpha, with special emphasis on the family Limoniidae. Acta Zool Cracoviensia. 35:11–36.
- Timmermans MJ, Vogler AP. 2012. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea). Mol Phylogenet Evol. 63(2):299–304.
- Wang Y, Liu X, Garzón-Orduña IJ, Winterton SL, Yan Y, Aspöck U, Aspöck H, Yang D. 2017. Mitochondrial phylogenomics illuminates the evolutionary history of Neuropterida. Cladistics. 33(6):617–636.
- Wang Y, Liu X, Winterton SL, Yang D. 2012. The first mitochondrial genome for the fishfly subfamily Chauliodinae and implications for the higher phylogeny of Megaloptera. PLOS One. 7(10):e47302.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Yang J, Yang D. 1988. Six new species of the genus Tanyptera from China (Diptera: Tipulidae). J Hubei Univ (Nat Sci). 10:70–75.
- Zhang X, Kang Z, Ding S, Wang Y, Borkent C, Saigusa T, Yang D. 2019. Mitochondrial genomes provide insights into the phylogeny of Culicomorpha (Insecta: Diptera). IJMS. 20(3):747.
- Zhang X, Kang Z, Mao M, Li X, Cameron SL, de Jong H, Wang M, Yang D. 2016. Comparative Mt genomics of the Tipuloidea (Diptera: Nematocera: Tipulomorpha) and its implications for the phylogeny of the Tipulomorpha. PLOS One. 11(6):e0158167.
- Zhao C, Qian X, Wang S, Li Y, Zhang X. 2019. The complete mitochondrial genome and phylogenetic analysis of Tipula (Yamatotipula) nova Walker, 1848 (Diptera, Tipulidae) from Qingdao, Shandong, China. Mitochondrial DNA Part B. 4(2):4211–4213.
