Abstract
Fenerivia ghesquiereana (Cavaco & Keraudren) R.M.K. Saunders (Annaonaceae) is a rare and endemic tree restricted to the warm tropical forests of Madagascar, a major global hotspot for biodiversity. Species in the genus are mostly restricted to a thin belt along the eastern edge of the island and remain under intense pressure due to deforestation for logging, mining and slash-and-burn agriculture (‘tavy’), despite national pledges to curb biodiversity losses and increase park protection to combat illegal logging and poaching. Here we report the complete chloroplast genome sequence of this species to support ongoing efforts to complete the (sub-)tribal classification of the family. The chloroplast sequence of F. ghesquiereana was 160,194 bp in length, including two inverted repeat regions of 26,093 bp, a large single-copy region of 89,041 bp and a small single-copy region of 18,967 bp. A total of 160 genes were annotated, of which 115 are coding, 37 are tRNA genes, and eight are rRNA genes. The overall GC content was 39%; this was higher in the IRs (43.4%) when compared to the LSC (30.7%) and the SSC (33.9%) regions. A Maximum Likelihood phylogenetic analysis with a selection of other plastomes in Annonaceae placed F. ghesquiereana as sister to Meiogyne hainanensis (Merr.) in subfamily Malmeoideae.
The Annonaceae are a diverse family of ca. 108 genera and 2400 species of trees, shrubs and lianas (Chatrou, Pirie, et al. Citation2012). They are among the most important components of tropical forests worldwide (Couvreur et al. Citation2018). Many species are also widely cultivated throughout the tropics and subtropics for their fleshy edible fruits, and for their spice, aromatic, and medicinal properties (Chatrou, Erkens, et al. Citation2012; Strijk et al. Citation2020).
The genus Fenerivia (∼10 spp.) is endemic to Madagascar (Saunders et al. Citation2011) and was included in the genus Polyalthia until recently. Morphological and phylogenetic studies (based on nuclear and chloroplast markers) have provided support for its recognition as an independent genus embedded in tribe Fenerivieae of the Malmeoideae subfamily (Deroin Citation2007; Saunders et al. Citation2011; Chatrou, Pirie, et al. Citation2012).
Fenerivia ghesquiereana (Cavaco & Keraudren) R.M.K. Saunders (vern. name ‘Korofoka’) is a rare tree confined to the wet tropical rain forests lining the eastern edge of Madagascar. Here, we report the complete chloroplast sequence of F. ghesquiereana (Cavaco & Keraudren) R.M.K. Saunders. To date, plastome data for the genus was not available and could not be included in any major molecular workup of the family. This new data will contribute to the delineation of relationships of Fenerivia with other genera in future studies.
We extracted genomic DNA from silica-dried leaves collected separately Madagascar (voucher ARI31, 14°40′48.5″S, 49°31′43.1″E) deposited in the TEF herbarium at Antananarivo, using a modified SDS protocol (Hinsinger & Strijk Citation2017). Library construction and sequencing were performed by Novogene (Beijing, China), using an Illumina HiSeq2500 platform following system manufacturer instructions. We conducted a de novo assembly of the chloroplast genome with Novoplasty v3.8.2 (Dierckxsens et al. Citation2017) using one GB of sequence data. The annotation was performed with GeSeq (Tillich et al. Citation2017) and was manually adjusted in Geneious (R9, Biomatters, www.geneious.com).
In F. ghesquiereana, the complete chloroplast genome (GenBank accession MT780300) was 160,194 bp in length, including two inverted repeat (IRS) regions of 26,093 bp, a large single-copy (LSC) region of 89,041 bp and a small single-copy (SSC) region of 18,967 bp. 160 genes were annotated, of which 115 were coding, 37 were tRNA genes, and eight were rRNA genes. The overall GC content was 39%. GC content was higher in the IRs (43.4%) than in LSC (30.7%) and SSC (33.9%) regions.
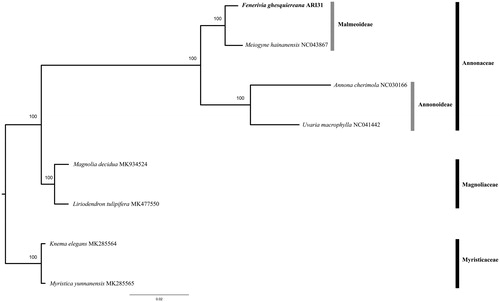
We explored phylogenetic relationships of F. ghesquiereana with other species in Annonaceae and outgroup species in Myristicaceae and Magnoliaceae, using additional plastomes available in GenBank (Annonaceae: Annona cherimola Mill. NC030166; Meiogyne hainanensis (Merr.) Tien Ban NC043867; Uvaria macrophylla Roxb. NC041442. Magnoliaceae: Magnolia decidua (Q.Y. Zheng) V.S. Kumar MK934524; Liriodendron tulipifera L. MK477550. Myristicaceae: Knema elegans Warb. MK285564; Myristica yunnanensis Y.H. Li MK285565).
The four main chloroplast regions (LSC, IRb, SSC and IRa) were automatically extracted, re-positioned and aligned using the software EcuADOR (Armijos Carrion et al. Citation2020) for all new and downloaded sequence data. A Maximum Likelihood phylogenetic tree was reconstructed using RaxML-NG (Kozlov et al. Citation2019) using 1000 bootstrap replicates. Fenerivia ghesquiereana was placed as sister to Meiogyne hainanensis in subfamily Malmeoideae with species in the Annonoideae subfamily as sister (Annona cherimola and Uvaria macrophylla) ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in GenBank (https://www.ncbi.nlm.nih.gov/) under accession number MT780300.
Additional information
Funding
References
- Armijos Carrion AD, Hinsinger DD, Strijk JS. 2020. ECuADOR-Easy Curation of Angiosperm Duplicated Organellar Regions, a tool for cleaning and curating plastomes assembled from next generation sequencing pipelines. PeerJ. 8:e8699.
- Chatrou LW, Erkens RH, Richardson JE, Saunders RMK, Fay MF. 2012. The natural history of Annonaceae. Botanical J Linn Soc. 169(1):1–4.
- Chatrou LW, Pirie MD, Erkens RH, Couvreur TL, Neubig KM, Abbott JR, Mols JB, Maas JW, Saunders RM, Chase MW. 2012. A new subfamilial and tribal classification of the pantropical flowering plant family Annonaceae informed by molecular phylogenetics. Bot J Linn Soci. 169(1):5–40.
- Couvreur TL, Helmstetter AJ, Koenen EJ, Bethune K, Brandão RD, Little SA, Sauquet H, Erkens RH. 2018. Phylogenomics of the major tropical plant family Annonaceae using targeted enrichment of nuclear genes. Front Plant Sci. 9:1941–1941.
- Deroin T. 2007. Floral vascular pattern of the endemic Malagasy genus Fenerivia Diels (Annonaceae). Adansonia. 29:7–12.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hinsinger DD, Strijk JS. 2017. Complete chloroplast genome sequence of Castanopsis concinna (Fagaceae), a threatened species from Hong Kong and South-Eastern China. Mitochondrial DNA A. 28(1):65–66.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Saunders RM, Su YC, Xue B. 2011. Phylogenetic affinities of Polyalthia species (Annonaceae) with columellar‐sulcate pollen: enlarging the Madagascan endemic genus Fenerivia. Taxon. 60(5):1407–1416.
- Strijk JS, Hinsinger DD, Roeder MM, Chatrou LW, Couvreur TL, Erkens RH, Sauquet H, Pirie MD, Thomas DC, Cao K. 2020. Chromosome-level reference genome of the Soursop (Annona muricata), a new resource for Magnoliid research and tropical pomology. Authorea.DOI: 10.22541/au.159103606.66673541
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.