Abstract
Huodendron tibeticum (J.Anthony) Rehder, which plays an important role in ecology and economy, is a deciduous species of Styracaceae. The authors sequenced, assembled, and annotated the chloroplast (cp) genome of Huodendron tibeticum using the sequencing data from Illumina Novaseq platform in this study. The complete cp genome of H. tibeticum is 159,320 bp in length, including a large single-copy (LSC) region of 87,795 bp, and a small single-copy (SSC) region of 18,989 bp. It contains 130 genes, including 37 tRNA genes, 8 rRNA genes, and 85 protein-coding genes. The overall GC content of H. tibeticum chloroplast genome is 36.66%. The phylogenetic analysis suggests that H. tibeticum is a sister species to H. biaristatum in Styracaceae.
Huodendron tibeticum (J.Anthony) Rehder is a prominent species of Styracaceae, 6–25 m tall trees or shrubs, and 25 cm diameter of trunk at breast height, which mainly distribute in dense forests at an altitude of 1000–3000 m in the west and southwest (northeast Guangxi, Guizhou, west Hunan, southeast Xizang, Yunnan) of China and Sino Indian Peninsula (Huang and Grimes Citation2003). It is valued for its timber, ornamental, perfume, and afforestation purposes. However, there has been little progress on its complete chloroplast genome. In this work, we characterized the complete cp genome sequence of H. tibeticum (GeneBank accession number: MT806103) based on Illumina pair-end sequencing data to provide a valuable complete cp genomic resource.
The fresh leaves of H. tibeticum were collected from Qingxiu Mountain (N 22.0780, E 108.3780) in Nanning, Guangxi, China. And total genome DNA was extracted using PlantDNA Kit (Genepioneer Biotechnologies, Nanjing, China). The herbarium of Nanjing Forestry University has deposited the voucher specimen (accession number 2020092). Using ultrasound to break DNA and the fragments of DNA were passivated, repaired, and bonded. The DNA fragments were selected by agarose gel electrophoresis. The sample of genome sequencing library was formed by PCR amplification, which was carried out on Illumina Novaseq platform by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China), and read long for PE150 sequencing.
The original reading was filtered by fastp (version 0.20.0), and the clean data were assembled into chloroplast genome using SPAdes (Bankevich et al. Citation2012). Next, the reference sequence (Genebank accession number: NC041127.1) was used for quality control after assembly. Finally, the assembled genome was annotated using CpGAVAS (Liu et al. Citation2012) and the physical map of H. tibeticum cp genome was drawn by the OGDRAW (Greiner et al. Citation2019). Based on the Maximum Likelihood (ML), the phylogenetic tree was concluded by MAFFT (Katoh et al. Citation2019) and MEGA (version 7) (Kumar et al. Citation2016).
The complete chloroplast genome sequence of H. tibeticum was 159,320 bp in length. The genome had a typical quadripartite structure including a pair of IR (IRa and IRb) regions of 26,268 bp that were separated by an LSC region of 87,795 bp and a SSC region of 18,989 bp. A total of 130 genes were encoded, including 8 rRNA genes (4 rRNA species), 37 tRNA genes (30 tRNA species), and 85 protein-coding genes (79 CDS species). Most of the genes occurred in a single copy; however, six protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7 and ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC), and four distinct rRNA gene (23S, 16S, 5S and 4.5S) are duplicated. A total of 10 protein-coding genes (accD, atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16) contained 1 intron while the other 2 genes (rsp12, ycf3) had 2 intron each. The overall GC content of the chloroplast genome is 36.66%. In addition, the GC contents of the LSC, SSC and IR regions are 34.74%, 30.26% and 42.18%, respectively.
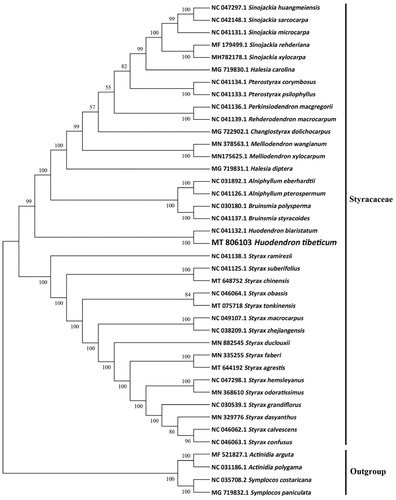
To reveal the phylogenetic evolution of H. tibeticum, we constructed a ML phylogenetic tree based on 36 cp genomes from Styracaceae and 4 cp genomes as outgroups from 2 taxa (Actinidiaceae, Symplocaceae). We found that H. tibeticum was clustered with other families of Styracaceae with 100% boot-strap values (). What’s more, H. tibeticum was highly supported to be a sister species to Huodendron biaristatum in Styracaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data are accessible from https://pan.baidu.com/s/1gcgDnY3I1nzzLYpcRUiOow (password: b7na); https://pan.baidu.com/s/1IRQxW8SP8gV7PK6Spz0xrg (password:1kn4).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Huang S-M, Grimes JW. 2003. Styracaceae. In: Wu Z.-Y., Raven, P.H. & Hong D.-Y. (Eds.) Flora of China, Vol. 15 (Styracaceae). Beijing: Science Press; St. Louis, MO: Missouri Botanic Garden Press; p. 265.
- Katoh K, Rozewicki J, Yamada KD. 2019. Mafft online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715