Abstract
The complete chloroplast genome sequence of Anemone reflexa exhibited a typical quadripartite structure with two inverted repeats (IRa and IRb) of 31,260 bp, a large single-copy region (LSC) of 80,767 bp, and a small single-copy region (SSC) of 17,623 bp. The chloroplast genome encoded a set of 133 genes, comprised of 89 protein coding genes, 36 tRNA genes and 8 rRNA genes. Phylogenetic analysis suggested that A. reflexa was closely related to A. raddeana. The complete chloroplast genome sequence of A. reflexa will provide valuable genetic resources for molecular identification and phylogenetic analysis of Anemone.
The Anemone genus, belonging to the family Ranunculaceae, has long been used in folk medicine and worldwide ethnomedicine, and more than 50 species in this genus have ethnopharmacological uses (Hao et al. Citation2017). Anemone reflexa is distributed in China, Korea, Mongolia and Siberia, and usually grows in open forests and valleys (Wang and Ziman Citation2001). Due to morphological similarity, rhizomes of A. reflexa are usually collected together with other traditional herbal medicines such as that of A. altaica and A. raddeana in the local market. The whole plastomes can be used as super-barcodes for accurate classification and identification for closely related species (Li et al. Citation2015; Fu et al. Citation2019). In the present study, the complete chloroplast genome sequence of A. reflexa is reported contributing to the molecular identification and phylogenetic analysis of Anemone.
The fresh and healthy leaves of an individual of A. reflexa in Longyuwan, Luanchuan county, Henan province (111°47′50.80″E, 33°40′50.68″N, 1733 m) were collected and stored in plastic bags with silica gel until DNA extraction. The specimen was deposited at Zhengzhou University (accession number: LY2019041111). Total genomic DNA was extracted according to a modified CTAB method (Doyle and Doyle Citation1987) and the shotgun library with an average insert size of 500 bp fragments was constructed. High-throughput DNA sequencing was conducted using the NovaSeq6000 System with S1 FlowCell (Illumina Inc., San Diego, CA) with 150 bp paired-end reads in Sangon Biotech (Shanghai) Co., Ltd. Approximately 3.3 Gb raw data were generated and used to assemble the chloroplast genome by GetOrganelle (Jin et al. Citation2020).Genome annotation was performed by GeSeq (Tillich et al. Citation2017) with A. raddeana as the reference and checked manually using Geneious Prime (Kearse et al. Citation2012). The annotated chloroplast genome of A. reflexa was submitted to GenBank with accession number MW043774. The complete chloroplast genome of A. reflexa and other eleven species from Ranunculaceae were aligned with MAFFT (Katoh and Standley Citation2013). Take Actaea dahurica as the outgroup, the maximum likelihood (ML) tree was constructed by RAxML (Stamatakis Citation2014) based on rapid bootstrap algorithm with the GTRGAMMA model and 1000 bootstrap replications.
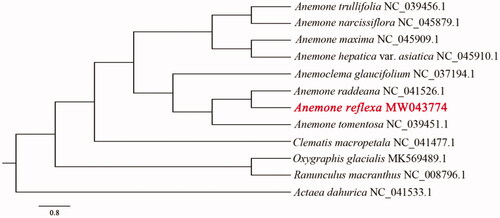
The complete chloroplast genome of A. reflexa is 1,60,910 bp in length, exhibiting a typical quadripartite structure with two inverted repeats (IRa and IRb: 31,260 bp). The large single-copy region (LSC) and the small single-copy region (SSC) separated by IRs are 80,767 bp and 17,623 bp, respectively. Totally, 133 genes were annotated, including 89 protein coding genes, 36 tRNA genes and 8 rRNA genes. The GC content of the whole genome, IRs, LSC and SSC regions are 37.64%, 41.90%, 35.78% and 31.08%, respectively. Phylogenetic analysis results indicate A. reflexa is closely related to A. raddeana (), which is congruent with previous molecular results (Hoot et al. Citation2012; Zhai et al. Citation2019).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that newly obtained at this study are openly available in the NCBI (https://www.ncbi.nlm.nih.gov/) under accession number of MW043774.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fu CN, Wu CS, Ye LJ, Mo ZQ, Liu J, Chang YW, Li DZ, Chaw SM, Gao LM. 2019. Prevalence of isomeric plastomes and effectiveness of plastome super-barcodes in yews (Taxus) worldwide. Sci Rep. 9(1):2773.
- Hao DC, Gu X, Xiao P. 2017. Anemone medicinal plants: ethnopharmacology, phytochemistry and biology. Acta Pharm Sin B. 7(2):146–158.
- Hoot SB, Meyer KM, Manning JC. 2012. Phylogeny and reclassification of Anemone (Ranunculaceae), with an emphasis on austral species. Syst Bot. 37(1):139–152.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. 2015. Plant DNA barcoding: from gene to genome. Biol Rev Camb Philos Soc. 90(1):157–166.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45(W1):W6–W11.
- Wang WC, Ziman SN. 2001. Ranunculaceae. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 6. Beijing: Science Press and Missouri Botanical Garden Press; p. 323–324.
- Zhai W, Duan XS, Zhang R, Guo CC, Li L, Xu GX, Shan HY, Kong HZ, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21.