Abstract
The Chinese endemic species Prunus hypoxantha is of great importance biogeographically, and is distributed in the edge of the Qinghai-Tibetan Plateau. Here, the complete chloroplast genome sequence of P. hypoxantha was assembled based on the Illumina reads. The complete plastome is 158,740 bp in length, with a large single-copy (87,206 bp) region, a small single-copy (18,884 bp) region, and two inverted repeat (26,325 bp) regions. The plastome contains 134 genes. Complete chloroplast genome sequence of P. hypoxantha will provide irreplaceable information in rebuilding the evolutionary history of the Maddenia clade.
Prunus hypoxantha (Koehne) J. Wen is a member of the Maddenia clade of Prunus. It grows in alpine forests of western to central China (Kalkman Citation2004; Wen and Shi Citation2012). As its distribution area includes the east most edge of the Qinghai-Tibetan Plateau (QTP), its phylogenetic research may be helpful to explain the speciation on QTP. Here, the complete plastid genome of P. hypoxantha (GenBank accession number: MK911761) was assembled by using the genome skimming approach (Zimmer and Wen Citation2015).
Fresh leaves of P. hypoxantha was sampled from Caiyuanzigou, Kangding Country, Sichuan Province, China (101°57’47′’E, 30°03’15′’N, alt. 2705 m), where the type specimen was collected, and dried with silica gel. A voucher specimen (ZL201708002) was deposited in the Herbarium of Northwest A&F University (WUK), China. Total genomic DNA was extracted with CTAB method (Doyle and Doyle Citation1987). A 500-bp DNA TruSeq Illumina (Illumina Inc., San Diego, CA, USA) sequencing library was constructed and then sequenced on Illumina Miseq platform (Illumina, San Diego, CA, USA). Reads of the chloroplast genome were assembled with GetOrganelle (Jin et al. Citation2020). The complete cp genome was annotated using PGA (https://github.com/quxiaojian/PGA) and corrected with DOGMA (Wyman et al. Citation2004). We submitted all the newly sequence data in raw format (fastq) and with the SRA accession (SRR12920654).
The complete plastome sequence of P. hypoxantha is 158,740 bp in length. The length of each inverted repeat (IR) is 26,325 bp, while the length of the large single-copy (LSC) and the short single-copy (SSC) is 87,206 bp and 18,884 bp, respectively. 134 genes were found in this plastome, 87 of them are unique protein-coding genes, while 39 are unique tRNA genes and 8 are unique rRNA genes. Most of the genes have a single copy, while nine tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnR-CCG, trnN-GUU and trnG-GCC), 5 protein-coding genes (rpl2, rpl23, ycf2, ndhB and rps7) and all the four rRNA genes are duplicated. The protein-coding gene rpl2 has three copies. The overall G/C content in the plastome is 36.6%.
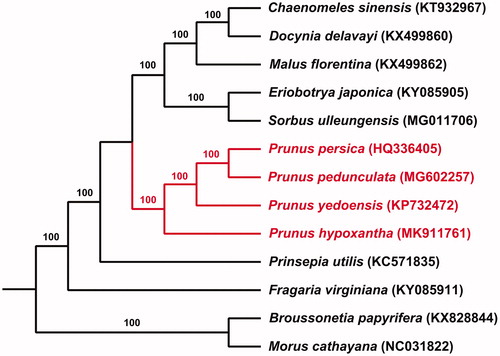
Totally 13 species (including 11 of Rosaceae, and Broussonetia papyrifera and Morus cathayana of Moraceae as outgroups) were used as outgroups to obtain the phylogenetic position of P. hypoxantha by RAxML. P. hypoxantha as well as the four other Prunus species forms a clade, which supports that P. hypoxantha, once belongs to a single genus Maddenia, should be a member of Prunus (). Such result was consistent with several other molecular studies (Wen et al. Citation2008). The complete chloroplast sequence of P. hypoxantha will provide useful resource for further phylogenetic and biogeographic study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov](https://www.ncbi.nlm.nih.gov/) under the accession no. MK911761. The associated BioProject, SRA, and Bio-Sample numbers are PRJN 671803, S12920654, and S16552112 respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241.
- Kalkman K. 2004. The families and genera of vascular plants. Berlin: Springer. p. 343–386.
- Wen J, Berggren ST, Lee CH, Ickert-Bond S, Yi TS, Yoo KO, Xie L, Shaw J, Potter D. 2008. Phylogenetic inferences in Prunus (Rosaceae) using chloroplast ndhF and nuclear ribosomal ITS sequences. J Syst Evol. 46(3):322–332.
- Wen J, Shi WT. 2012. Revision of the Maddenia clade of Prunus (Rosaceae). PhytoKeys. 11:39–59.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zimmer EA, Wen J. 2015. Using nuclear gene data for plant phylogenetics: progress and prospects II. Next-gen approaches. J Syst Evol. 53(5):371–379.