Abstract
Lysimachia christinae Hance is widely distributed in subtropical China at the elevational range from 500–2300 m. The species is an important medicinal herb for treating jaundice, urinary disorders, and the liver. Here, we sequenced and characterized the whole plastid genome of L. christinae. It is 154,810 bp in length, containing two copies of inverted repeat (IR) regions (26,034 bp, each), a large single-copy (LSC) region (84,809 bp), and a small single-copy (SSC) region (17,933 bp). It has 114 genes, of which 80 are protein-coding, 30 are tRNA, and 4 are rRNA genes. The ML tree indicates L. christinae is closely related to Lysimachia congestiflora Hemsl. This genome information can help us better construct a backbone phylogeny of Lysimachia in the future.
Lysimachia L. is a large genus in the northern hemisphere in the family Primulaceae, containing 211 described species, with ∼81% occurring in East Asia (Yan et al. Citation2018). Lysimachia christinae Hance is widely distributed in the subtropical region in China growing at the elevational range from 500–2300 m (Hu and Kelso Citation1996). The species is a famous medicinal herb that is widely used to treat jaundice, urinary disorders, and the liver (Chen and Hu Citation1979). In this study, we characterized the complete plastid genome of L. christinae to determine its genome structure and evolutionary relationship to other Primulaceae species.
Fresh leaves of an individual of L. christinae, collected from Gulin County of Sichuan Province (28°16′01′′N, 105°49′39′′E), were used for DNA extraction with a modified CTAB protocol (Doyle and Doyle Citation1987). The voucher specimen (no. Y2011046) was deposited at the Herbarium of South China Botanical Garden. The Illumina paired-end (PE = 150 bp) library was constructed and sequenced on the Illumina Hiseq X Ten platform at the Beijing Genomics Institute (Wuhan, China). Low-quality reads and adaptor sequences were removed from ∼2 G bp raw data. The plastome of L. christinae was assembled using NOVOPlasty (Dierckxsens et al. Citation2017) with the reference plastome (NC_026197) of Lysimachia coreana Nakai (Son and Park Citation2016). The draft plastid genome was checked by remapping the clean reads in Geneious Prime 2019 (Biomatters, Ltd, Auckland, New Zealand). The tRNA genes were annotated using ARAGORN (Laslett and Canback Citation2004) and the coding genes with DOGMA (Wyman et al. Citation2004). Both genes were manually adjusted using Geneious Prime 2019. Other plastid genomes from the Primulaceae were downloaded from GenBank for the phylogenetic analyses. The aligned matrix comprised of 79 shared protein-coding genes extracted from 27 whole plastid genomes (including the newly generated plastome herein) was implemented in MAFFT (Katoh and Standley Citation2013), and used for ML tree construction. The maximum-likelihood (ML) tree was constructed using RAxML (Stamatakis Citation2014) with 1000 rapid bootstrap replicates.
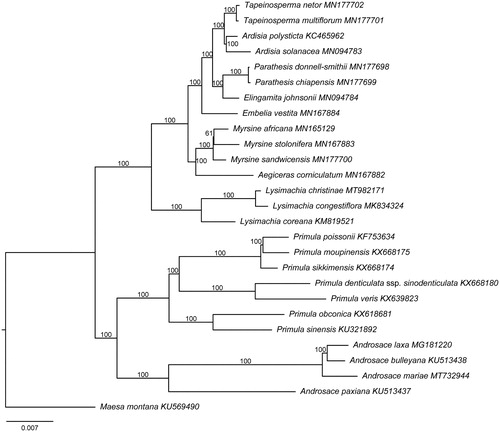
The plastid genome of L. christinae (GenBank accession number MT982171) is 154,810 bp in length with a typical quadripartite structure. It contains two copies of inverted repeat (IR) regions (26,034 bp), a large single-copy (LSC) region (84,809 bp), and a small single-copy (SSC) region (17,933 bp). A total of 114 unique genes were encoded, of which 80 are protein-coding genes, 30 are tRNA genes, and four are rRNA genes. The overall GC content of the genome is 37%. Specifically, the GC values of the LSC, SSC, and IR regions are 34.8, 30.4, and 42.8%, respectively. The plastid genome structures of all three Lysimachia species are highly conserved since no genome rearrangement was detected (Son and Park Citation2016; Li et al. Citation2019). However, Lysimachia coreana (belonging to the subgen. Palladia) has only 79 protein-coding genes in its plastome, while both from the subgen. Lysimachia – Lysimachia congestiflora Hemsl. and L. christinae – contain 80 protein-coding genes (Li et al. Citation2019). The ML tree showed the species of Lysimachia formed a monophyletic clade with 100% bootstrap support value (). Lysimachia congestiflora and L. christinae were recovered in a sister relationship with full support (bootstrap value = 100%), both belonging to the subgen. Lysimachia (Yan et al. Citation2018). The robust sister relationship between the subgen. Lysimachia and the subgen. Palladia was confirmed in this study, in agreement with the results of Anderberg et al. (Citation2007) and Yan et al. (Citation2018).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/, reference number MT982171. The raw sequencing data is available under GenBank Bioproject PRJNA670596.
Additional information
Funding
References
- Anderberg AA, Manns U, Källersjö M. 2007. Phylogeny and floral evolution of the Lysimachieae (Ericales, Myrsinaceae): evidence from ndhF sequence data. Willdenowia. 37(2):407–421.
- Chen FH, Hu CM. 1979. Taxonomic and phytogeographic studies on Chinese species of Lysimachia. Acta Phytotaxon Sin. 17:21–56.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hu CM, Kelso S. 1996. Primulaceae. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 15. Beijing (China): Science Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Li H-L, Cheng X-L, Chen Y, Tan F-L. 2019. Complete plastome sequence of Lysimachia congestiflora Hemsl. a medicinal and ornamental species in Southern China. Mitochondrial DNA Part B. 4(2):2316–2317.
- Son O, Park SJ. 2016. Complete chloroplast genome sequence of Lysimachia coreana (Primulaceae). Mitochondrial DNA Part A. 27(3):2263–2265.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yan HF, Zhang CY, Anderberg AA, Hao G, Ge XJ, Wiens JJ. 2018. What explains high plant richness in East Asia? Time and diversification in the tribe Lysimachieae (Primulaceae). New Phytol. 219(1):436–448.