Abstract
Ficus altissima plays an important role on biodiversity in tropical forests. In this study, the complete chloroplast genome sequence and the genome features of F. altissima were analyzed using the Illumina NovaSeq platform. The whole chloroplast genome sequence of F. altissima is 160,251 including a large single-copy region (LSC, 88,468 bp), a small single-copy region (SSC, 20,009 bp), and a pair of repeat regions (IRs, 25,887 bp, each). Further gene annotation revealed the chloroplast genome contains 124 genes, including 79 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. A total of 82 simple sequence repeats (SSRs) were identified in the chloroplast genome. Phylogenetic development was analyzed based on F. altissima with other species of Moraceae. This information will be useful for study on the evolution and genetic diversity of F. altissima in the future.
It is widely accepted that tropical rainforests support the richest biodiversity among terrestrial ecosystems. However, they are suffering from rapid loss of biodiversity due to lots of reasons, such as human disturbance and climate change (Ghazoul and Sheil Citation2010). As a keystone resource in tropical forests, Ficus L. constitutes the most distinctive of the widespread genera in tropical area since figs (syconium) have a complex obligatory mutualism with their pollinating agaonid fig wasps (Weiblen Citation2002; Dunn Citation2020), and figs are found in almost all tropical habitat types and geographic locations (Janzen Citation1979). Furthermore, Ficus as a genus comprising more than 800 species, also playing a critical role in biodiversity conservation in the tropics, because Ficus are the most important group of plants for fruit-eating vertebrates (Shanahan et al. Citation2001).
Ficus altissima Blume (subgenus Urostigma) is a monoecious fig tree species distributed across Asia (Berg and Corner Citation2005). It occurs naturally in tropical forests at Xishuangbanna, Yunnan, China. F. altissima is also frequently planted in cities and villages or near temples as an ornamental or sacred plant. The aerial root and branch of F. altissima can be used as traditional Chinese medicine, and tender leaves are also used as edible vegetable by Xishuangbanna local people. In recent years, the ecology of F. altissima has intrigued the interests of ecologists (Peng et al. Citation2008, Citation2010; Zhang et al. Citation2014), while the complete chloroplast genome has not been sequenced. Considering the chloroplast DNA-based studies can provide invaluable data for studying genetic history and phylogeny, and can also provide important information in designing conservation strategies for the species, in this study, we sampled the F. altissima from the Xishuangbanna tropical botanical garden, Chinese Academy of Science (21°55′ N, 101°15′ E, 555 m above sea level). A voucher specimen (YAB 202,005) was deposited at Yunnan Academy of Biodiversity, Southwest Forestry University, Yunnan, China. Then we sequenced, assembled and annotated the accurate chloroplast genome with the next-generation sequencing method to reveal the phylogenetic relationship of F. altissima.
For this study, the total genomic DNA of F. altissima was extracted from fresh leaves according to the modified CTAB methods described by Doyle and Doyle (Citation1987). A genomic shotgun library with an insertion size of 340 bp was constructed. The libraries were constructed using standard protocols (NEB Next Ultra II DNA Library Prep Kit for Illumina), and sequenced on Illumina NovaSeq platform at Personalbio Biotech (Shanghai, China). We assembled the chloroplast genome using GetOrganelle software version 1.7.1 (Jin et al. Citation2020), and the assembled chloroplast genome was annotated through the online program CPGAVAS 2 (Shi et al. Citation2019) with F. religiosa chloroplast genome (GenBank accession number: NC_033979) as a reference, and assisted with manual correction. The raw sequencing reads used in this study has been deposited in SRA with the accession number SRR12665077 and the annotated chloroplast genome sequence has been deposited into the GenBank with the accession number MW013819.
The complete chloroplast genome of F. altissima was 160,251 bp in length, consisting of a large single copy region (LSC, 88,468 bp), a small single copy region (SSC, 20,009 bp), and two inverted repeat regions (IRa and IRb, 25, 887 bp). The total GC content was 36.92%, with IR regions (42.60%) higher than that in LSC (33.56%) and SSC regions (28.9%). The chloroplast genome of F. altissima contains a total of 124 genes, including 79 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. Furthermore, a total of 82 SSR markers ranging from mononucleotide to pentanucleotide repeat motif were identified in F. altissima chloroplast genome.
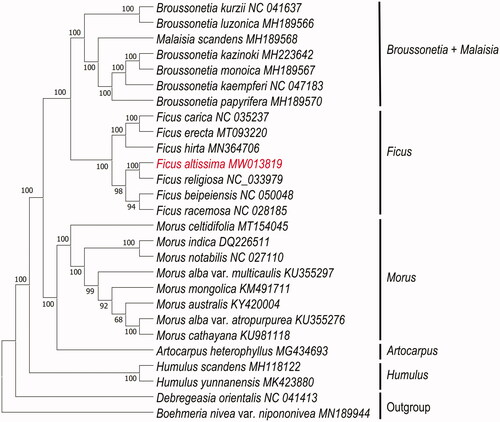
To determine the phylogenetic relationship of F. altissima, based on complete chloroplast genomes of 25 species within the family Moraceae, with Debregeasia orientalis and Boehmeria nivea var. nipononivea as outgroup (), chloroplast genomes were downloaded from NCBI. All chloroplast genomes were aligned using the program MAFFT v7.471 (Rozewicki et al. Citation2019), and phylogenetic tree (maximum likelihood) constructed by Iqtree software version 1.6.12 (Minh et al. Citation2020) with 1000 bootstrap replicates, best-fitted model has been confirmed is TVM + F+R2 by ModelTest-NG (Darriba et al. Citation2020). The phylogenetic analysis revealed that F. altissima closely clustered with F. religiosa. The complete chloroplast genome of F. altissima will provide essential data for future research on the phylogenetic and evolutionary relationship in genus of Ficus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the analyses and results of this study are openly available in Genbank with accession (MW013819) (https://www.ncbi.nlm.nih.gov/). Raw sequencing reads was deposited in SRA with BioProject accession (PRJNA664225). (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA664225).
Additional information
Funding
References
- Berg CC, Corner EJH. 2005. Moraceae (Ficus). In Nooteboom HD. Leiden: National Herbarium of Nederland. p. 1–625.
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Dunn DW. 2020. Stability in fig tree-fig wasp mutualisms: how to be a cooperative fig wasp. Biol J Linn Soc. 130(1):1–17.
- Ghazoul J, Sheil D. 2010. Tropical rain forest ecology, diversity, and conservation. Oxford: Oxford University Press.
- Janzen DH. 1979. How to be a fig. Annu Rev Ecol Syst. 10(1):13–51.
- Jin JJ, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. Getorganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Peng YQ, Duan ZB, Yang DR, Rasplus JY. 2008. Co-occurrence of two Eupristina species on Ficus altissima in Xishuangbanna, Southwestern China. Symbiosis. 45:9–14.
- Peng YQ, Yang DR, Compton SG. 2010. The reproductive success of Ficus altissima and its pollinator in a strongly seasonal environment: Xishuangbanna, Southwestern China. Plant Ecol. 209(2):227–236.
- Rozewicki J, Li SL, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10.
- Shanahan M, So S, Compton SG, Corlett R. 2001. Fig-eating by vertebrate frugivores: a global review. Biol Rev Camb Philos Soc. 76(4):529–572.
- Shi LC, Chen HM, Jiang M, Wang LQ, Xi W, Huang LF, Chang L. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Weiblen GD. 2002. How to be a fig wasp. Annu Rev Entomol. 47:299–330.
- Zhang Y, Peng YQ, Compton SG, Yang DR. 2014. Premature attraction of pollinators to inaccessible figs of Ficus altissima: a search for ecological and evolutionary consequences. PLos One. 9(1):e86735.