Abstract
The complete chloroplast genome of Sibiraea angustata was assembled and subjected to phylogenetic analysis. The chloroplast genome of S. angustata was 155,869 bp in length, containing a large single-copy region (84,343 bp), a small single-copy region (18,820 bp), and two inverted repeat regions (26,353 bp). The overall GC content of S. angustata chloroplast genome was 36.80%. The chloroplast genome of S. angustata contained 127 unique genes, including 83 protein-coding genes, 36 tRNA genes and eight rRNA genes. Phylogenetic analysis revealed that S. angustata was related to Malus ioensis, Malus florentina and Malus trilobata.
Sibiraea angustata (Rehd.) Hand.-Mazz. is a typical plant in the Qinghai-Tibet Plateau, mainly distributed in the shrub of 3000–4000 m in Western China (Wu Citation1998). The twigs and fresh leaves of S. angustata are used as traditional Chinese medicine to treat stomach discomfort and indigestion (Liu et al. Citation2020). To make better use of S. angustata, the complete chloroplast genome of S. angustata was assembled and subjected to phylogenetic analysis.
Fresh leaves of S. angustata were sampled from Ruoergai County, Aba State, Sichuan Province, China (N33°37′45.55″, E102°36′20.14″). The specimen was deposited in the Laboratory of Forest Pathology, College of Forestry, Sichuan Agricultural University (Voucher No. SAHM-202005177). Total genomic DNA was extracted according to the mCTAB protocol (Li et al. Citation2013). The genome sequencing was conducted using Illumina NovaSeq platform by Shanghai Personal Biotechnology Co. Ltd, China. The chloroplast genome of S. angustata was assembled with SPAdes v3.6.0 software (Bankevich et al. Citation2012) and annotated with Plann (Huang and Cronk Citation2015). The complete chloroplast genome sequence of S. angustata has been submitted to the GenBank (MW123094).
The chloroplast genome of S. angustata was 155,869 bp in length, containing a large single-copy (LSC) region (84,343 bp), a small single-copy (SSC) region (18,820 bp), and two inverted repeat (IR) regions (26,353 bp). The overall GC content of S. angustata chloroplast genome was 36.80%, and in the SSC, LSC, and IR regions were 30.64%, 34.61%, and 42.51%, respectively. The chloroplast genome contained 127 complete genes, including 8 rRNA genes, 36 tRNA genes, and 83 protein-coding genes.
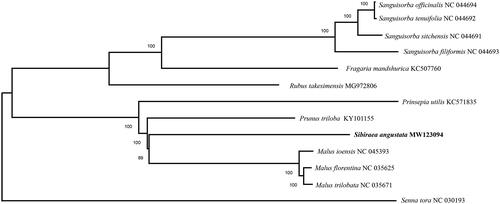
To evaluate the phylogenetic relationships of S. angustata within Rosaceae, the chloroplast genome sequence of S. angustata and those of 12 other Rosaceae species were aligned by MAFFT v7 (Katoh and Standley Citation2013), and the phylogenetic tree was constructed with MEGA X using neighbour-joining analysis (Kumar et al. Citation2018). The phylogenetic tree revealed that S. angustata was most related with Malus ioensis, Malus florentina and Malus trilobata ().
Date availability statements
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number MW123094. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA669189, SUB8684720, and SAMN16457089 respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li J, Wang S, Jing Y, Wang L, Zhou S. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48(1):72–78.
- Liu H, Zhang T, Jiang P, Zhu W, Yu S, Liu Y, Li B, Li F. 2020. Hypolipidemic constituents from the aerial portion of Sibiraea angustata. Bioorg Med Chem Lett. 30(11):127161.
- Wu L. 1998. The community types and biomass of Sibiraea angustata scrub and their relationship with environmental factors in northwestern Sichuan. Acta Bot Sin. 40(9):860–870.