Abstract
Alloteropsis is a widely-distributed genus with C3 and C4 photosynthetic species belonging to the Poaceae family. This study characterized the complete chloroplast genomes of two Alloteropsis species from Chinese mainland, i.e. Alloteropsis semialata with C4 photosynthetic type and Alloteropsis cimicina with C3 photosynthetic type. The chloroplast genomes of A. semialata and A. cimicina are 138,188 bp and 139,335 bp in length, with 38.48% and 38.59% GC contents, respectively. These two chloroplast genomes contain the same number of genes, i.e. 107 unique genes including 27 tRNA, 4 rRNA, and 76 protein-coding genes. Phylogenomic analysis confirmed the species identity of A. semialata and A. cimicina and supported a close relationship of Alloteropsis with species of Setaria and Panicum in grass family.
Alloteropsis (Poaceae: Paniceae) is a genus that comprises approximately 5 species, including C3 and C4 photosynthetic types (Hattersley and Watson Citation1992). This genus has a wide geographical distribution, occurring in tropical and Southern Africa, India, SE Asia and Australia. There are two Alloteropsis species in China, i.e. A. semialata (R. Brown) Hitchcock and A. cimicina (Linnaeus) Stapf (Chen and Phillips Citation2006). Alloteropsis semialata is the only known grass which has both C3 and C4 photosynthetic forms (Ibrahim et al. Citation2009) while its sister species A. cimicina is C3 plant. In 1974, Ellis firstly reported that there were two different anatomical structures within A. semialata, i.e. ‘Kranz anatomy’ structure and ‘non-Kranz anatomy’ structure (Ellis Citation1974). According to the leaf anatomical structure and the different photosynthetic types, this species has been divided into two subspecies: A. semialata subsp. semialata (C4 type) and A. semialata subsp. eckloniana (C3 type) (Russell Citation1983). So far, according to our survey, only C4 photosynthetic type A. semialata is collected in China and the other species of the same genus, A. cimicina has C3 photosynthetic type. Alloteropsis has been an intriguing model to study the evolution of C4 photosynthesis. However, those are insufficient genomic data for molecular study of this genus, compared to other cereal crops such as maize and sorghum (Wang et al. Citation2009). On this account, this study generated the complete chloroplast genomes of A. semialata and A. cimicina from Chinese mainland, and it has complemented the data of Alloteropsis species in Chinese mainland and coupled with the latest assembly and annotation software.
Young, fresh, and healthy leaves were collected from A. semialata in Yunnan (26°27′38″N, 99°53′43″E) and A. cimicina in Hainan (19°29′58″N, 110°13′54″E). Both voucher specimens were deposited in the herbarium of Kunming Institute of Botany, Chinese Academy of Science (KUN) with accession numbers of YY3-JC-20 and YY14-Ac-HN-4, respectively. Genomic DNA was extracted via CTAB method (Doyle Citation1987), then prepared and sequenced on the Illumina Hiseq 4000 platform. About five Gb paired-end data were produced for each species and the data were assembled using GetOrganelle (Jin et al. Citation2020). To determine the accuracy, we mapped the reads to the result of GetOrganelle in Geneious version 9.1.4 (Kearse et al. Citation2012). The assembled cp genome was annotated using PGA (Qu et al. Citation2019), coupled with manual check and adjustment. The ecotype RCH20 of A. cimicina (NC_027952) was used as reference for assembling and annotation.
The complete chloroplast genome sequence of A. semialata (GenBank accession number MT950759) is 138,188 bp in length. The large single-copy (LSC) and small single-copy (SSC) regions are 81,946 bp and 12,618 bp, which are separated by a pair of inverted repeats (IRs) with 21,812 bp for each. The GC content of whole genome is 38.48%. This genome contains 107 unique genes, including 76 protein-coding genes, 27 tRNA genes, and 4 rRNA genes.
The complete chloroplast genome sequence of A. cimicina (GenBank accession number MT950760) is 139,335 bp in length. The lengths of LSC, SSC and IR are 81,747 bp, 12,688 bp and 22,450 bp, respectively. The genome GC content is 38.59%. There are also 107 unique genes in this genome, including 76 protein-coding genes, 27 tRNA genes, and 4 rRNA genes.
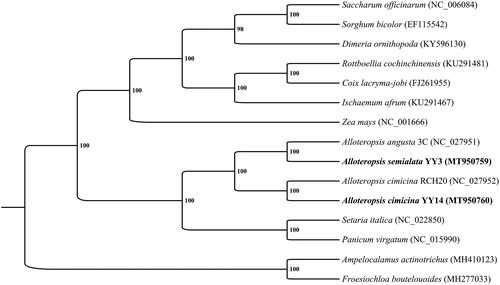
To confirm the phylogenetic location of A. semialata and A. cimicina within the family of Poaceae, a total of 15 chloroplast genomes from Poaceae were analyzed. RAxML–HPC BlackBox (Stamatakis Citation2014) was used for estimating the maximum likelihood (ML) tree through Cipres Science Gateway (Miller et al. Citation2010). The result () confirmed the species identity of A. semialata and A. cimicina samples in this study, which clustered to C3 ecotype of A. angusta and RCH20 ecotype A. cimicina with high supported values (100%), respectively. A close relationship among Alloteropsis, Setaria and Panicum was supported too. These newly sequenced chloroplast genomes of Alloteropsis would facilitate to study of the evolution of C3 and C4 photosynthetic forms in the genus Alloteropsis.
Figure 1. Phylogenetic tree produced by maximum likelihood (ML) analysis base on 15 chloroplast genome sequences from Poaceae, including the newly sequenced A. semialata and A. cimicina and their closely-related species (the remaining 13 chloroplast genome sequences were download from NCBI, and the GenBank Number is in the parentheses behide every species name, respectively). YY3 is the abbreviation for accession numbers of YY3-JC-20, and YY14 is the abbreviation for accession numbers of YY14-Ac-HN-4. Numbers associated with branched are assessed by Maximum Likelihood bootstrap support values.
Acknowledgments
We appreciate the Laboratory of Molecular Biology in the Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, Chinese Academy of Sciences for providing experimental platform.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT950759- MT950760. The associated’BioProject’, ‘SRA’ and ‘Bio-Sample’ numbers are PRJNA672776- PRJNA672775, SRR12927465- SRR12927463, and SAMN16576581- SAMN16576580 respectively.
Additional information
Funding
References
- Chen SL, Phillips SM. 2006. Alloteropsis (Poaceae). In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; Vol. 22; p. 519–520.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Ellis RP. 1974. The significance of occurrence of both Kranz and non-Kranz leaf anatomy in the grass species Alloteropsis semialata. S Afr J Sci. 70(6):169–173.
- Hattersley PW, Watson L. 1992. Diversification of photosynthesis. In: Chapman GP, editor. Grass evolution and domestication. Cambridge (UK): Cambridge University Press; p. 38–116.
- Ibrahim DG, Burke T, Ripley BS, Osborne CP. 2009. A molecular phylogeny of the genus Alloteropsis (Panicoideae, Poaceae) suggests an evolutionary reversion from C4 to C3 photosynthesis. Ann Bot. 103(1):127–136.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Paper presented at: Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14; New Orleans (LA); p. 1–8.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Russell GEG. 1983. The taxonomic position of C3 and C4 Alloteropsis semialata (Poaceae) in southern Africa. Bothalia. 14(2):205–213.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang XY, Gowik U, Tang HB, Bowers JE, Westhoff P, Paterson AH. 2009. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 10(6):R68.
