Abstract
Bryophyllum daigremontianum is a very important traditional medicine and ornamental plant. Although Bryophyllum and Kalanchoe have been supported to form a clade, however, lack of chloroplast genomic severely hinders our understanding the phylogenetic relationships between them. In this study, the complete chloroplast genome of B. daigremontianum is first presented. It is 150,058 bp in length consisted a large single-copy (LSC, 82,164 bp) and a small single-copy (SSC, 17,042bp) separated by a pair of inverted repeats (IR, 25,426 bp) including 86 protein-coding genes, 37 tRNA, and 8 rRNA. Phylogenetic analysis supported that B. daigremontianum was closer to K. tomentosa than other species, which showed that chloroplast genome sequences offer a useful resource for future phylogenetic studies of Kalanchoe and Bryophyllum species.
Bryophyllum daigremontianum (Raym.-Hamet & Perrier) A. Berger is well known for its interesting reproduction and important traditional medicine distributed in tropics, such as South Asia, South Africa, and Australia (Bernard Citation2006; Milad et al. Citation2014; Kolodziejczyk-Czepas and Stochmal Citation2017; Stefanowicz-Hajduk et al. Citation2020). It is closely related to the genus Kalanchoe based on the morphology and molecular phylogenetic analysis, such as matK, ITS (Mort et al. Citation2001, Citation2005). Previous studies also found the species could hybrids between Bryophyllum Berger and Kalanchoe Adans., which makes the boundary of species between these two genera more confused (Hart and Eggli Citation1996; Van Ham and Hart Citation1998).
In recent years, chloroplast genomes have important contributions to phylogenetic studies of plants (Daniell et al. Citation2016). However, few chloroplast genomic data are available for these genera, only one species of Kalanchoe in GenBank database, K. tomentosa Baker (Accession number: MN794319). As part of the phylogenetic study to understand the relationship between Bryophyllum and Kalanchoe, we reported the whole chloroplast genome of B. daigremontianum and its phylogenetic analysis with related species.
Sample of B. daigremontianum was collected from Zhengzhou, Henan province, China (34°76′N, 119°72′E), specimen in the herbarium of Zhengzhou University (no. ZZU2020-201). High-quality genomic DNA was extracted from fresh leaves using Tiangen Plant Genomic DNA Kits (TIANGEN, Beijing) and directly constructed short-insert of 150 bp in length libraries and sequenced on the Illumina Genome Analyzer (Hiseq 2500) based on the manufacturer’s protocol by Novogene Inc., Beijing. De novo assembly was done using CLC Genomics Workbench v11.0 (Qiagen Inc., Aarhus, Denmark) and Geneious R11 (Kearse et al. Citation2012) with reference of K. tomentosa. Finally, the sequence was annotated by PGA (Qu et al. Citation2019) and Geseq online program (Tillich et al. Citation2017).
Approximately 4.0 G raw data were assembled of B. daigremontiana chloroplast genome sequence with length 150,058 bp, which submitted to GenBank with the accession number MT954417. The cp genome was showed a typical quadripartite structure, consisted of a large single-copy region with 82,164 bp (LSC) and small single-copy region with 17,042 bp (SSC), separated by a pair of inverted repeat regions with 25,426 bp (IRs), each. The whole genome sequence GC content is 37.62%, while in LSC, SSC and IR regions are 35.63%, 31.39% and 42.92%, respectively. A total of 131 genes were predicted in the whole chloroplast genome of B. daigremontianum, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
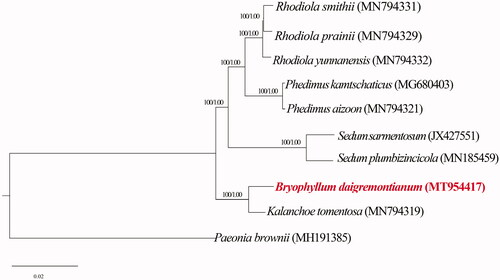
The phylogenetic analysis of B. daigremontiana and other eight species of Crassulaceae were based on Bayesian inference using Mybayes v3.2.7 (Ronquist et al. Citation2012) and maximum likelihood (ML) method using RaxMLGUI v2.0 (Silvestro and Michalak Citation2012) with Paeonia brownii Dougl. ex Hook (Accession number: MH191385) as outgroup. The tree was constructed using GTRGAMMA substitution model. From the result of two phylogenetic tree, B. daigremontianum was close to K. tomentosa formed a strongly supported clade (). These researches will provide useful information for the phylogenetic and evolutionary studies of the genus Bryophyllum and Kalanchoe in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MT954417. The associated SRA number is PRJNA668018.
Additional information
Funding
References
- Milad R, ElAhmady S, Singab AN. 2014. Genus Kalanchoe (Crassulaceae): a review of its ethnomedicinal, botanical, chemical and pharmacological properties. EJMP. 4(1):86–104.
- Bernard D. 2006. Le genre Kalanchoe (Crassulaceae): structure et définition. Le Journal de Botanique France. 33:29–32.
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134.
- Hart H, Eggli U. 1996. Evolution and systematics of the Crassulaceae. British Cactus Succulent Soc. 14:32.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kolodziejczyk-Czepas J, Stochmal A. 2017. Bufadienolides of Kalanchoe species: an overview of chemical structure, biological activity and prospects for pharmacological use. Phytochem Rev. 16(6):1155–1171.
- Mort ME, Levsen N, Randle CP, Van Jaarsveld E, Palmer A. 2005. Phylogenetics and diversification of Cotyledon (Crassulaceae) inferred from nuclear and chloroplast DNA sequence data. Am J Bot. 92(7):1170–1176.
- Mort ME, Soltis DE, Soltis PS, Francisco-Ortega J, Santos-Guerra A. 2001. Phylogenetic relationships and evolution of Crassulaceae inferred from matK sequence data. Am J Bot. 88(1):76–91.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MRBAYES 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Stefanowicz-Hajduk J, Asztemborska M, Krauze-Baranowska M, Godlewska S, Gucwa M, Moniuszko-Szajwaj B, Stochmal A, Ochocka JR. 2020. Identification of flavonoids and bufadienolides and cytotoxic effects of Kalanchoe daigremontiana extracts on human cancer cell lines. Planta Med. 86(4):239–246.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Van Ham R, Hart H. 1998. Phylogenetic relationships in the Crassulaceae inferred from chloroplast DNA restriction-site variation. Am J Bot. 85(1):123.