Abstract
Saccostrea echinata is a rock perched oyster with wide distribution and tremendous potential for commercial mariculture. However, the taxonomy of this genus is confused. In this study, we described the complete mitochondrial genome of medium-sized form of Saccostrea echinata. The genome is 16,282 bp in length, encoding the standard set of 12 protein-coding genes (PCGs), 23 tRNA genes, and two rRNA genes, with circular organization. The overall base composition of the whole mitochondrial genome was A (26.78%), T (36.64%), G (21.99%), and C (14.59%) with an AT bias of 63.42%. The longest PCG of these species was ND5, whereas the shortest was ND4L.
The giant or blacklip oyster Saccostrea echinata (Quoy and Gaimard 1835) that live on rocky shores are broadly distributed throughout the western and south Pacific. It has the commercial potential of mariculture within the Asia-Pacific region due to the suitability of this species for hatchery culture (Glude Citation1984; Southgate and Peter Citation1998; Angell et al. Citation2009). Species in genus Saccostrea are well known for the highly morphological plasticity with different habitat conditions that morphological diagnosis are extremely difficult and time-consuming (Lam and Morton Citation2006; Amaral and Luiz Citation2016). DNA-sequenced based methods provide an opportunity to construct additional taxon diagnosis systems that employ DNA sequences as ‘barcodes’ to identify oyster and to conduct further phylogenetic studies (Sekino and Yamashita Citation2016; Volatiana et al. Citation2016; Cui Citation2018; Samantha et al. Citation2019). Here, we sequenced and annotated mitogenome of S. echinata which will provide essential molecular information for the genetical understanding of this oyster and thus contribute to better management and sustainable use of the species.
The specimens of S. echinata are collected from Dajin island, northwest of South China Sea (21°52′N, 113°2′E) in 16 September 2020. Whole genomic DNA was extracted from muscle tissue of one specimen of S. echinata using TIANamp Marine Animals DNA Kit (TIANGEN, Beijing, China). The concentration for use as a PCR template was adjusted to an A260 of about 0.05–0.2. The collected specimen and extracted DNA were stored in Guangdong Provincial Key Laboratory of Fishery Ecology and Environment (specimen accession number: JMDJD2020-C3).
DNA sequencing was carried out using genomic DNA by ABI 3730xl DNA automatic sequencer with PCR primers designed from highly conserved regions of transfer RNA (tRNA) sequences of related species (Wu et al. Citation2019) with additional specific primers designed as required from sequences already obtained. The COI sequence of S. echinata was used as reference seeds for iterative assembly by MITObim v.1.8 (Hahn et al. Citation2013). SeqMan v.7.1.0 was used for the mitogenome assembly and annotation (Swindell and Plasterer Citation1997). Transfer RNA genes were predicted using online software tRNAScan-SE 1.21 (Lowe and Eddy Citation1997). All protein-coding gene (PCG) are aligned independently, then concatenated to be applied for phylogenetic reconstruction with other Bivalvia species in MrBayes v 3.12 (Ronquist and Huelsenbeck Citation2003) using relaxed clock model.
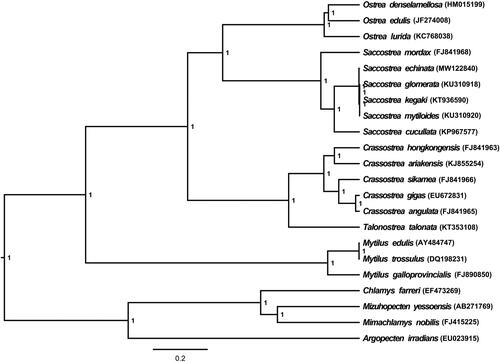
S. echinata (MW122840) mitochondrial genome forms a 16,282 bp closed loop. The overall base composition of the whole mitochondrial genome was A (26.78%), T (36.64%), G (21.99%), and C (14.59%) with an AT bias of 63.42%. This mitochondrial genome represents a typical Ostreidae mitochondrial genome and matches with the Saccostrea kegaki (KT936590) genome, in which it comprises 12 protein coding gene, 23 tRNA genes, and two rRNA genes (12S rRNA and 16S rRNA) and one A + T-rich region which could also be termed as control region. The 16S rRNA gene is split into two parts by a large fragment. The ATG and GTG initiation codons are used in most of PCGs except ND41 and CYTB (ATA), and the stop codons of all the 12 PCGs were complete. Six PCGs (COX1, CYTB, COX2, ND1, ND4, and ND4L) use TAA as the termination codon; six PCGs (ND3, ND5, ND6, COX3, ATP6, and ND2) use TAG as the termination codon. Meanwhile, the longest PCG of these species was ND5 (1671 bp), whereas the shortest ND4L (281 bp). All the 21 typical tRNAs possess a complete clover leaf secondary structure, ranging from 61 bp to 71 bp. In order to understand the evolutionary relationship and position of S. echinata, a Bayesian inference phylogenetic tree was constructed based on mitochondrial PCGs and rRNAs of 21 other Bivalvia species, of which 14 species belong to the same family (Ostreidae) as S. echinata and five belong to genus Saccostrea. The Bayesian inference phylogenetic tree showed that S. echinata (MW122840) firstly grouped with S. glomerata (KU310918), S. kegaki (KT936590), and S. mytiloides (KU310920), then closely related to S. cucullate (KP967577) (). Support values of Bayesian Posterior were showed on the branches. Phylogenetic analyses consistent with the classification results depended on the morphological characters. We have the confidence to construct phylogenetic trees, based on the complete mitochondrial genomes, but the evolution history of Ostreidae still needs future research to be clearly resolved.
Disclosure statement
None of the coauthors has any conflict of interest to declare. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov reference number MW122840. The authors confirm that the data supporting the findings of this study are available at the following link: https://doi.org/10.5281/zenodo.4271298.
Additional information
Funding
References
- Amaral VSD, Luiz RLS. 2016. Comparative anatomy of five species of Saccostrea Dollfus and Dautzenberg, 1920 (Bivalvia: Ostreidae) from the Pacific Ocean. Nautilus. 130(2):53–71.
- Angell CL, Tetelepta J, Smith LS. 2009. Culturing the spiny oyster, Saccostrea echinata, in Ambon, Indonesia. J World Aquacul Soc. 15(1–4):433–441.
- Cui XM. 2018. Taxonomic and phylogenetic studies of Saccostrea oysters on the coastal areas of China [Master thesis]. Qingdao: Chinese Academy of Sciences Institution of Oceanology; pp. 2–5.
- Glude JB. 1984. The applicability of recent innovations to mollusc culture in the western Pacific islands. Aquaculture. 39(1–4):29–43.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Lam K, Morton B. 2006. Morphological and mitochondrial DNA analysis of the Indo-West Pacific rock oysters (Ostreidae: Saccostrea species). J Mollus Stud. 72(3):235–245.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Samantha JN, Catarina NSS, Paul CS, Jan MS. 2019. Mitochondrial and nuclear genetic analyses of the tropical black-lip rock oyster (Saccostrea echinata) reveals population subdivision and informs sustainable aquaculture development. BMC Genomics. 20:711.
- Sekino M, Yamashita H. 2016. Mitochondrial and nuclear DNA analyses of Saccostrea oysters in Japan highlight the confused taxonomy of the genus. J Mollus Stud. 82(4):492–506.
- Southgate PC, Peter SL. 1998. Hatchery rearing of the tropical blacklip oyster Saccostrea echinata Quoy and Gaimard. Aquaculture. 169(3–4):275–281.
- Swindell SR, Plasterer TN. 1997. Seqman, contig assembly. In: Swindell SR, editor. Sequence data analysis guidebook. Totowa (NJ): Springer; p. 75–89.
- Volatiana JA, Fang SS, Kinaro ZO, Liu X. 2016. Complete mitochondrial DNA sequences of Saccostrea mordax and Saccostrea cucullata: genome organization and phylogeny analysis. Mitochondrial DNA Part A. 27(4):3024–3025.
- Wu JH, Ju YM, Hsiao ST. 2019. The complete mitochondrial genome of Saccostrea kegaki (Pterioida, Ostreidae). Mitochondrial DNA Part B. 4(1):642–643.