Abstract
Torreya parvifolia (Torreya, Taxaceae) is endemic in Sichuan, China. It consisted of an extremely small population with less than 100 wild individuals. In this study, the complete chloroplast genome of T. parvifolia was assembled using the Illumina data. The complete chloroplast genome of T. parvifolia is 137,106 bp in length. The genome consists of 119 genes in total, including 82 protein-coding genes (PCGs), 4 ribosomal RNA (rRNA) genes, and 33 transfer RNA (tRNA) genes. Phylogenetic analysis indicated that T. parvifolia was closely related to T. fargesii, T. nucifera, and T. fargesii var. yunnanensis with strong support.
Torreya parvifolia T.P. Yi, Lin Yang & T.L. Long, belonging to Torreya in Taxaceae family, is a species with extremely small population (Yi et al. Citation2006). It is reported that there are less than 100 individuals surviving in the wild (Pan et al. Citation2014). In this study, we reported the complete chloroplast genome of T. parvifolia for the first time and performed a phylogenetic analysis with 11 other species based on their complete chloroplast genomes.
We collected the fresh leaves of a wild T. parvifolia individual from Liangshan Yi Autonomous Prefectu, Sichuan Province, China (27°30′N, 102°56′E). Voucher specimen of the species was deposited in the Ecological Security and Protection Key Laboratory of Sichuan Province, China under the accession number: MNU-PHO-0160. The total DNA was extracted with the CTAB method (Doyle and Doyle Citation1987). We performed the whole-genome sequencing with HiSeq2500 Platform (Illumina, San Diego, CA) and obtained ∼10 Gb high-quality clean data. The complete chloroplast genome of T. parvifolia was de novo assembled with NOVOPlasty (Dierckxsens et al. Citation2017). The gene prediction was carried out by Plann (Huang and Cronk Citation2015) and Sequin (NCBI website). Finally, we obtained a chloroplast genome of T. parvifolia. The genome has been submitted to the GenBank under the accession number of NC_043866.1.
The complete chloroplast genome of T. parvifolia is 137,106 bp in length, with a GC content of 35.47% in total. The genome structure is similar to other Taxaceae species (Miu et al. Citation2018; Ge et al. Citation2019; Shin et al. Citation2019), with the loss of one copy of the inverted repeats (IRs). The chloroplast genome of T. parvifolia contains 119 genes, including 82 protein-coding genes (PCGs), 4 ribosomal RNA (rRNA) genes, and 33 transfer RNA (tRNA) genes.
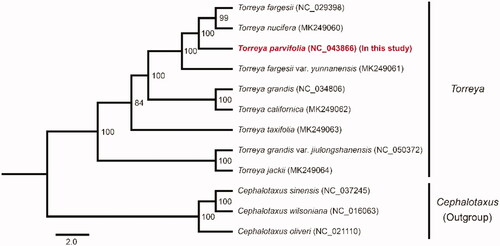
To infer the phylogenetic position of T. parvifolia, we reconstructed a phylogenetic tree using the concatenated 64 PCGs sequences of T. parvifolia and 11 other species. The sequences of each gene were aligned by PRANK (Löytynoja Citation2014). RAxML (Stamatakis Citation2014) was performed to construct the phylogenetic relationships with 100 bootstrap replicates under the GTRGAMMA model. The maximum likelihood (ML) tree revealed T. parvifolia was closely related to T. fargesii, T. nucifera, and T. fargesii var. yunnanensis with strong support (). Within the concatenated 64 PCGs sequences, T. parvifolia had 50, 187, and 162 varied sites with T. fargesii, T. nucifera, and T. fargesii var. yunnanensis, respectively. The genetic relationships of other Torreya species were identical with the previous study (Zhang et al. Citation2019) (). Furthermore, we found T. fargesii did not cluster with T. fargesii var. yunnanensis. T. grandis and T. grandis var. jiulongshanensis were also not clustered together. It was possible because of the mis-identification of materials for sequencing, or the inaccurate published new varieties, which were only based on the characteristics of morphology in past years. In summary, the chloroplast genome of T. parvifolia could help us facilitate the identification and protection for T. parvifolia.
Disclosure statement
No potential competing interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. NC_043866.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA683944, SRR13235763, and SAMN17048565, respectively. The voucher specimen of the species is free accessible at Ecological Security and Protection Key Laboratory of Sichuan Province, China with the no. MNU-PHO-0160.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Ge J, Bi G, Luo G. 2019. Characterization of complete chloroplast genome of Amentotaxus yunnanensis (Taxaceae), a species with extremely small populations in Chin. Mitochondrial DNA. 4(1):1765–1767.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Löytynoja A. 2014. Phylogeny-aware alignment with PRANK. In: Russell D, editor. Multiple sequence alignment methods, methods in molecular biology. Totowa (NJ): Humana Press; p. 155–170.
- Miu ZP, Zhang JM, Li JH, Hong X, Pan T. 2018. The complete chloroplast genome sequence of a conifer plant Torreya grandis (Pinales, Taxaceae). Mitochondrial DNA. 3(2):1152–1153.
- Pan H, Feng Q, Long T, He F, Liu X. 2014. Discussion on resource condition and protection technique for rare endangered species in Sichuan Province. J Sichuan Forestry Sci Technol. 35(6):41–46.
- Shin S, Kim SC, Hong KN, Kang H, Lee JW. 2019. The complete chloroplast genome of Torreya nucifera (Taxaceae) and phylogenetic analysis. Mitochondrial DNA. 4(2):2537–2538.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yi T, Yang L, Long T. 2006. Torreya parvifolia, a new species of the Taxaceae from Sichuan, China. Bull Bot Res. 26(5):513–515.
- Zhang X, Zhang HJ, Landis JB, Deng T, Meng AP, Sun H, Peng YS, Wang HC, Sun YX. 2019. Plastome phylogenomic analysis of Torreya (Taxaceae). J Syt Evol. 57(6):607–615.