Abstract
The African armyworm, Spodoptera exempta, is an episodic migratory crop pest with an expanding distribution worldwide. This is the first report of the circular mitochondrial genome of S. exempta, with a length of 15,457 bp and an A + T content of 81.7%. It encoded a common set of 37 genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes, and contained a putative control region of 379 bp (94.7% in A + T proportion). The maximum-likelihood phylogenetic tree based on the complete mitogenome demonstrated that five species belonging to the Spodoptera genus formed one clade, in which S. exempta was the most isolated branch, followed by Spodoptera exigua. This data will contribute for the identification and phylogenetic analyses of S. exempta, providing useful information for its comprehensive control.
The African armyworm, Spodoptera exempta (Walker, 1856) (Lepidoptera: Noctuidae), is an episodic crop pest and the most potentially serious hazard among migratory pests in Africa (Brown and Swaine, 1966; Pringle Citation1980; Redman et al. Citation2010). Mass oviposition of S. exempta results in the initiation of larval outbreaks, which can cause severe damage to rangeland grasses and crops, including maize, millet, rice, wheat, among others (Parker and Gatehouse Citation1985a, Citation1985b; Xu et al. Citation2020). To date, S. exempta has been widely distributed worldwide, including Africa, Asia, North America, and Oceania (Gunn and Gatehouse Citation1985; Haggis Citation1986). In the last two years, the fall armyworm, Spodoptera frugiperda, has invaded India and most Southeast Asian countries, as well as China (Zhang et al. Citation2020). As a species from the same genus, S. exempta has gained public attention again given its potential damaging effect for East Asian countries, including China, Japan, Korea. The species identification and correct differentiation of S. exempta are very important for accurate monitoring and early control. In the present study, the complete mitochondrial genome sequence of S. exempta was determined.
The S. exempta specimen used in this study was obtained from an inbred strain established from eggs collected in 2014 near Greytown, South Africa (29°03′ S, 30°36′ E). The genomic DNA of a male moth was extracted using the Qiagen Genomic DNA kit (Cat. no.13323, Qiagen) followed by purity assessment with a NanoDrop One UV-Vis spectrophotometer (Thermo Fisher Scientific). The voucher specimen’s genomic DNA (AGIS-SE-ZA-2014) deposited at the Agricultural Genomics Institute in Shenzhen, China. A total of 0.5 μg of genomic DNA was used to construct a 350-bp insert library, which was then sequenced using the 150-bp paired-end mode in an Illumina NovaSeq 6000 system (San Diego, CA, USA). The mitogenome was assembled using the software NOVOPlasty v2.5.6 (Dierckxsens et al. Citation2017), and its annotation was conducted using MITOS2 (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013).
The complete mitogenome of S. exempta was a typical circular DNA molecule with 15,457 bp. The length was the longest compared with other four published Spodoptera species, which ranged from 15,365 to 15,388 bp (Wan et al. Citation2013; Wu et al. Citation2013; Seo et al. Citation2019). The mitogenome comprised 13 protein-coding genes (PCGs), 22 transfer RNA (tRNAs) genes, two ribosomal RNA genes, and one predicted control region. All five Spodoptera species, including S. exempta, shared the same gene order and possessed the ancestral gene order with trnM-trnI-trnQ of lepidopteran mitogenomes (Cao et al. Citation2012). The A + T content of the mitogenome was 81.7%, which was similar to that of other Spodoptera species (80.9 ∼ 81.3%) and fell within the range of the A + T content for other Lepidoptera species (Kim et al. Citation2006; Salvato et al. Citation2008). The start codon of all PCGs was ATN (where N represents A, T, or G), except for cox1 that begun with CGA. Most PCGs were terminated with TAA or TAG as the stop codon, with the exception of cox2 and nad4 that had the incomplete stop codon T, which could produce the functional stop codon TAA by the addition of 3’A residues in polyadenylation processes (Ojala et al. Citation1981). The 379-bp control region of S. exempta was located between srRNA and trnM, which are known to be involved in the initiation of transcription and/or replication in other insects (Zhang and Hewitt Citation1997). All tRNAs harbored typical predicted secondary cloverleaf structures, except trnS1 and trnY.
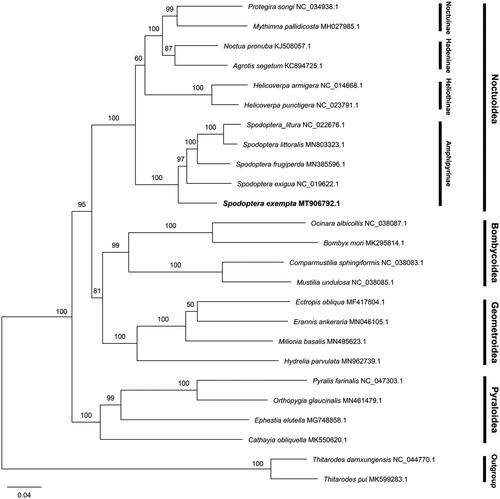
In order to determine the evolutionary relationship of S. exempta, a total of 25 complete mitogenomes (including 23 species in Heteroneura and 2 outgroup species from Exoporia) were used in phylogenetic analysis. 25 complete mitogenome sequences were multiple-aligned using MAFFT v7.455 (Katoh and Standley Citation2013). The phylogenetic tree was constructed based on the maximum likelihood method using RAxML-NG v0.9.0 (Kozlov et al. Citation2019) with the best-fit model (GTR + F+R4) estimated by IQ-TREE v1.6.10 with the parameter ‘-m MF’ (Nguyen et al. Citation2015). The bootstrap replicates were 1,000 and the tree was visualized with FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). The phylogenetic tree confirmed that S. exempta belongs to the Spodoptera genus with a high nodal supporting value, presenting the species relationships ([([S. littoralis + S. litura] + S. frugiperda) + S. exigua] + S. exempta) (). Different families of Lepidoptera and subfamilies of Noctuidae form one clade. The present data will contribute for the identification and phylogenetic assessment of S. exempta, and may provide useful information for the comprehensive control of hazardous crop pests.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MT906792. Associated BioProject, BioSample and SRA accession numbers are PRJNA678819, SAMN16814351, and SRR13083393, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Brown E, Swaine G. 1966. New evidence on the migration of moths of the African armyworm, Spodoptera exempta (Wlk.) (Lepidoptera, Noctuidae). Bull Entomol Res. 56(4):671–684.
- Cao YQ, Ma C, Chen JY, Yang DR. 2012. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics. 13:276
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Gunn A, Gatehouse AG. 1985. Effects of the availability of food and water on reproduction in the African army worm, Spodoptera exempta. Physiol Entomol. 10(1):53–63.
- Haggis M. 1986. Distribution of the African armyworm, Spodoptera exempta (walker) (Lepidoptera: Noctuidae), and the frequency of larval outbreaks in Africa and Arabia. Bull Entomol Res. 76(1):151–170.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim I, Lee EM, Seol KY, Yun EY, Lee YB, Hwang JS, Jin BR. 2006. The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Mol Biol. 15(2):217–225.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
- Parker WE, Gatehouse AG. 1985a. Genetic factors controlling flight performance and migration in the African armyworm moth, Spodoptera exempta (Walker) (Lepidoptera: Noctuidae). Bull Entomol Res. 75(1):49–64.
- Parker WE, Gatehouse AG. 1985b. The effect of larval rearing conditions on flight performance in females of the African armyworm, Spodoptera exempta (Walker) (Lepidoptera: Noctuidae). Bull Entomol Res. 75(1):35–48.
- Pringle JWS. 1980. Science of a pest: research on the African armyworm at the international center of insect physiology and ecology, Nairobi. Insect Biology in the Future. 925–945.
- Redman EM, Wilson K, Grzywacz D, Cory JS. 2010. High levels of genetic diversity in Spodoptera exempta NPV from Tanzania. J Invertebr Pathol. 105(2):190–193.
- Salvato P, Simonato M, Battisti A, Negrisolo E. 2008. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics. 9:331.
- Seo BY, Lee GS, Park J, Xi H, Lee H, Lee J, Park J, Lee W. 2019. The complete mitochondrial genome of the fall armyworm, Spodoptera frugiperda Smith, 1797 (Lepidoptera; Noctuidae), firstly collected in Korea. Mitochondrial DNA Part B. 4(2):3918–3920.
- Wan X, Kim MJ, Kim I. 2013. Description of new mitochondrial genomes (Spodoptera litura, Noctuoidea and Cnaphalocrocis medinalis, Pyraloidea) and phylogenetic reconstruction of Lepidoptera with the comment on optimization schemes. Mol Biol Rep. 40(11):6333–6349.
- Wu QL, Gong YJ, Gu Y, Wei SJ. 2013. The complete mitochondrial genome of the beet armyworm Spodoptera exigua (Hübner) (Lepodiptera: Noctuidae). Mitochondrial DNA. 24(1):31–33.
- Xu P, Yang L, Yang X, Li T, Graham RI, Wu K, Wilson K. 2020. Novel partiti-like viruses are conditional mutualistic symbionts in their normal lepidopteran host, African armyworm, but parasitic in a novel host, Fall armyworm. PLoS Pathog. 16(6):e1008467.
- Zhang DX, Hewitt GM. 1997. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 25(2):99–120.
- Zhang L, Liu B, Zheng W, Liu C, Zhang D, Zhao S, Li Z, Xu P, Wilson K, Withers A, et al. 2020. Genetic structure and insecticide resistance characteristics of fall armyworm populations invading China. Mol Ecol Resour. 20(6):1682–1696.