Abstract
The complete mitochondrial genome of Thelephora aurantiotincta, an edible Basidiomycete mushroom species with ecological and economic value is reported in this study. The whole genome is a circular molecule 50,672 bp in length and encodes 42 genes as follows: 15 protein-coding genes, two rRNA genes and 25 tRNA genes. The A, T, C, G contents in the genome are 35.60%, 35.31%, 13.89%, and 15.20%, respectively. Phylogenetic analysis revealed a close relationship between T. aurantiotincta and T. ganbajun. This is the first complete mitochondrial genome for T. aurantiotincta that will be useful for providing basic genetic information for this important species.
Thelephora species (Aphyllophorales, Thelephoraceae) usually form ectomycorrhizal associations with trees (Yagame and Maekawa Citation2019) and contribute significantly to plant health and ecosystem stability (Ramírez-López et al. Citation2015). Thelephora aurantiotincta Corner 1968 is a very common Thelephora species with a wide distribution in the Yunnan province of China. It is mainly distributed in braodleaf-conifer forest and coniferous forest, and harvested as a healthy and edible mushroom for its unique flavor (Norikura et al. Citation2011; Li Citation2016). In this study, we assembled and characterized the complete mitochondrial genome of T. aurantiotincta for the first time in order to provide genetic information for this edible mushroom. The annotated mitochondrial genome sequence was submitted to GenBank under the accession number of MT234668.
The sample of T. aurantiotincta was purchased in the Yunnan Mushuihua wild edible mushroom market in Kunming, China (N25°0′, E102°43′). Total genomic DNA was extracted and stored in Yunnan Agricultural University, Yunnan, China (contact person SC Shao, and email [email protected]) with the archival number of MG58. The DNA was sequenced on the Illumina Hiseq 4000 sequencing platform by Shanghai Personal Biotechnology Co. Ltd, China. Clean pair-end reads were filtered and assembled using SPAdes v.3.11.0 software (Bankevich et al. Citation2012) to yield the complete mitochondrial genome. Genes were annotated using the online GeSeq-Annotation of Organellar Genomes program (Tillich et al. Citation2017) (https://chlorobox.mpimp-golm.mpg.de/geseq.html) based on the T. ganbajun mitogenome (KY245891). The tRNA structural predictions were made using RNAstructure Web Servers for RNA Secondary Structure Prediction (http://rna.urmc.rochester.edu/RNAstructureWeb/) and tRNAscan-SE v2.0.5 (Lowe and Chan Citation2016).
The complete mitochondrial genome of T. aurantiotincta is 50,672 bp in length with the A, T, C, G contents of 35.60%, 35.31%, 13.89%, and 15.20%, respectively. A total of 42 genes were annotated, including 15 protein-coding genes, two rRNA genes and 25 tRNA genes. Three of the tRNA genes (trnI, trnL, trnF) are duplicated, and two (trnM and trnS) occur in triplicate in the mitochondrial genome. One protein-coding gene, cox1, has an intron. All the tRNA genes can be folded into classical cloverleaf secondary structures. The genetic compositions and coding sequences are similar to other mushroom species in Thelephora (Wang et al. Citation2017).
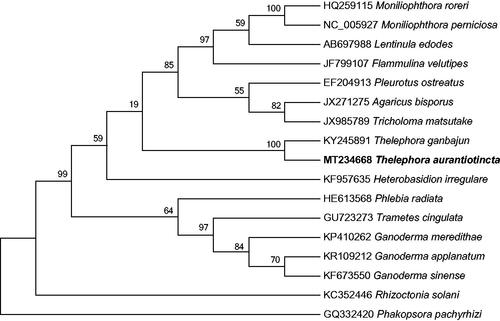
The phylogenetic relationships of T. aurantiotincta and sixteen other Agaricomycetes genomes were evaluated based on protein-coding genes. The phylogenetic tree was constructed using 1000 non-parametric bootstrap replicates with the maximum-likelihood analysis in MEGA 7.0 (Kumar et al. Citation2016) designating Phakopsora pachyrhizi as the outgroup taxon. The analysis found that the mitochondrial genome of T. aurantiotincta was fully resolved in a clade with T. ganbajun (). This is the first time to report the genetic relationship between Thelephora species at the molecular level, and the newly reported mitochondrial genome of T. aurantiotincta will provide valuable genetic information for species evolution and phylogenetic analysis of Thelephora and Thelephoraceae.
Figure 1. Phylogenetic relationship of Thelephora aurantiotincta and sixteen related species based on mitochondrial genome sequences. The phylogenetic tree was constructed using maximum-likelihood method based on the protein-coding genes, with Phakopsora pachyrhizi as the outgroup. The branch support was determined by computing 1000 non-parametric bootstrap replicates.
Disclosure statement
No potential competing interest was reported by the authors.
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at nucleotide database, https://www.ncbi.nlm.nih.gov/nuccore/MT234668, Associated BioProject, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA392534, BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/SAMN07303038 and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/SRR5803911.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li YP. 2016. Chemical constituents of essentialoil from the fungus Thelephora aurantiotincta. Chem Nat Compd. 52(5):932–933.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Norikura T, Fujiwara K, Narita T, Yamaguchi S, Morinaga Y, Iwai K, Matsue H. 2011. Anticancer activities of thelephantin O and vialinin A isolated from Thelephora aurantiotincta. J Agric Food Chem. 59(13):6974–6979.
- Ramírez-López I, Villegas-Ríos M, Salas-Lizana R, Garibay-Orijel R, Alvarez-Manjarrez J. 2015. Thelephora versatilis and Thelephora pseudoversatilis: two new cryptic species with polymorphic basidiomes inhabiting tropical deciduous and sub-perennial forests of the Mexican Pacific coast. Mycologia. 107(2):346–358.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45(W1):W6–W11.
- Wang PF, Zhang Y, Sha T, Xu JP. 2017. The complete mitochondrial genome of the edible Basidiomycete mushroom Thelephora ganbajun. Mitochondrial DNA B. 2(1):103–105.
- Yagame T, Maekawa N. 2019. Ectomycorrhiza synthesis and basidiome formation of an orchid symbiont of the genus Thelephora on Quercus serrata. Mycoscience. 60(6):343–350.
