Abstract
Thalictrum foeniculaceum is a morphologically distinctive species in the genus with very reduced filiform compound leaves and big pinkish flowers occurring in dry slopes of northern China. Herein, we report the first complete chloroplast genome sequence of T. foeniculaceum. The chloroplast genome sequence was 155,923 bp in length, with large and small single-copy regions (LSC with 85,323 bp and SSC with 17,628 bp in length) separated by two inverted repeat regions (IRs with 26,486 bp). The total GC content was 38.3%. The complete plastome sequence contained 111 genes, including 77 protein-coding, 30 tRNA, and four rRNA genes. The phylogenetic analysis of Thalictrum based on the complete chloroplast genomes available online was also presented in this study.
Thalictrum is a large genus with about 200 species almost worldwide in the buttercup family (Ranunculaceae) (Tamura Citation1995). Thalictrum species are known as the Traditional Chinese Medicines (TCM): horsetail goldthread. Thalictrum species have monochlamydeous flower which are often not showy. However, several Chinese species have large pinkish or purple flowers, such as T. delavayi, T. grandiflorum, T. diffusiflorum, T. chelidonii, and T. foeniculaceum (Wang and Wang Citation1979) and showed horticultural value. Among them, T. foeniculaceum is the most distinctive one with linear leaflets presumably adaptive to dry environments. In order to better understand this species, we reported and characterized the first complete chloroplast genome of T. foeniculaceum using the next-generation sequencing method.
Fresh young leaves of T. foeniculaceum were collected from Yunju Temple, Fangshan Dist., Beijing (115.766°E, 39.609°N). Voucher specimens were deposited in the Herbarium of Beijing Forestry University (BJFC) (under collection numbers of L. Xie 202007001). Genomic DNAs were extracted using a genomic DNA extraction kit (Tiangen Biotech Co. Ltd., Beijing, China), and 2 × 150 bp pair-end sequencing was performed on an Illumina HiSeq 4000 platform at Novogene (http://www.novogene.com, China). We used Map function of Geneious version R11 (Biomatters, Ltd., Auckland, New Zealand, Kearse et al. Citation2012) to select chloroplast reads using published plastome sequence of Thalictrum as reference (He et al. Citation2019). Then, these chloroplast reads were de novo assembled with Geneious R11. Gaps were filled using Fine Tuning function of Geneious R11. The assembled chloroplast sequence was annotated using the Plastid Genome Annotator (PGA, Qu et al. Citation2019), and verified by Geneious R11.
The complete chloroplast genome sequence of T. foeniculaceum is 155,923 bp in length, with a large single-copy (LSC) region of 85,323 bp, a small single-copy (SSC) region of 17,628 bp, and two inverted repeats (IR) of 26,486 bp. The plastome contains 111 functional genes, including 77 protein-coding genes, 30 tRNA genes, and four rRNA genes. The total sequence GC content is 38.3%. Structural variations, such as gene inversions, transpositions, and IR expansion of this plastome sequence was consistent with other Thalictrum samples reported by previous studies (He et al. Citation2019; Zhai et al. Citation2019).
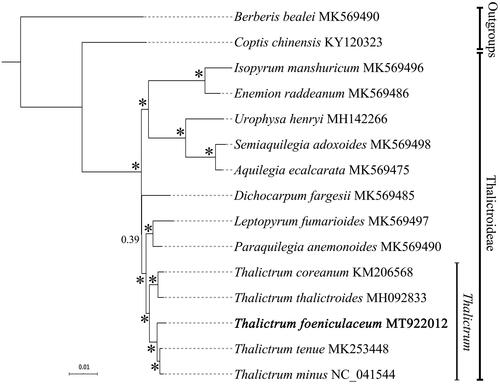
Complete chloroplast genome sequences of the other Thalictrum species available from GenBank were retrieved for phylogenetic analysis. Bayesian inference was used for phylogeny reconstruction (). The sequence alignment and all the settings of Bayesian analyses were followed (Liu et al. Citation2018). Phylogenetic framework of Thalictrum as well as outgroups were consistent with previous molecular studies (Soza et al. Citation2012; He et al. Citation2019; Zhai et al. Citation2019).
Disclosure statement
The authors declare that they have no conflict interests.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT922012. Raw Illumina data is available at the Sequence Read Archive (SRA) under accession SRR12718335.
Additional information
Funding
References
- He J, Yao M, Lyu RD, Lin LL, Liu HJ, Pei LY, Yan SX, Xie L, Cheng J. 2019. Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci Rep. 9(1):1–13.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu HJ, He J, Ding CH, Lyu RD, Pei LY, Cheng J, Xie L. 2018. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe Anemoneae (Ranunculaceae). Front Plant Sci. 9:1097.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Soza VL, Brunet J, Liston A, Salles Smith P, Di Stilio VS. 2012. Phylogenetic insights into the correlates of dioecy in meadow-rues (Thalictrum, Ranunculaceae). Mol Phylogenet Evol. 63(1):180–192.
- Tamura M. 1995. Ranunculaceae. In: Heipko P, editor. Engler’s die natürlichen pflanzenfamilien. 2nd ed. Berlin (Germany): Duncker & Humblot; p. 223–497.
- Wang WT, Wang SH. 1979. Thalictrum. In: Wu KF, editor. Flora reipublicae popularis sinicae. Vol. 13. Beijing (China): Science Press; p. 502–592.
- Zhai W, Duan XS, Zhang R, Guo CC, Li L, Xu GX, Shan HY, Kong HZ, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21.