Abstract
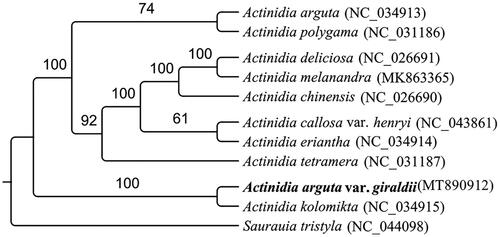
The complete chloroplast (cp) genome sequence of Actinidia arguta var. giraldii (Diels) Vorosch. was assembled and characterized by Illumina pair-end sequencing date. The complete plastid genome was 156,729 bp in length, containing a large single-copy (LSC) of 89,647 bp, and a small single-copy (SSC) of 22,482 bp, which was separated by a pair of 22,300 bp inverted repeat regions (IR). A total of 131 unique genes were identified, including 84 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. The phylogenetic position based on the chloroplast genome of 11 species showed that A. arguta var. giraldii was sister to Actinidia kolomikta.
Actinidia arguta is a climbing, perennial and dioecious vine, native from northeast Asian countries(China, Japan, Korea, and Siberia) (Zhu et al. Citation2019) and has become increasingly popular due to its exceptional flavor, interesting phytochemical profile, and outstanding pro-healthy properties (Latocha et al. Citation2015; Wojdyło and Nowicka Citation2019). Hardy kiwi is one of the most nutritionally rich fruits and an excellent source of antioxidants (mainly polyphenols), vitamins (especially vitamin C), carotenoids, chlorophylls, sugars, dietary fiber, organic acids, and minerals (Leontowicz et al. Citation2016; Latocha Citation2017; Diana et al. Citation2020). Hardy kiwi has been associated with the preventive effects of various chronic diseases, such as cardiovascular and digestive disorders. (Latocha et al. Citation2015; Latocha Citation2017). A. arguta var. giraldii (Diels) Vorosch. is a variety of A. arguta, mainly distributed in Shaanxi province in China, growing at an altitude of 1000 m. It produces small grape-sized fruits with thin, edible, and hairless skin and aromatic sweet flavor. These fruits come in green, yellow, or purple as it ripens which provides valuable materials to study the color of kiwi. The exploration of complete chloroplast genomes of it can provide a methodological guidance for evolution and phylogenetic analysis of Actinidia in the future.
The materials were introduced from Heilong River, Guyi District, Xi’an City, Shaanxi Province (108°46′E, 33°87′N) and the voucher specimens were stored at Xi’an Botanical Herbarium (XBGH). Total genomic DNA was extracted from young leaves using the modified CTAB method (Doyle Citation1987). Genome sequencing was performed using Illumina HiSeq 4000 platform at Biomarker Technologies Corporation (Beijing, China). The qualified cleaned reads were assembled with NOVOPlasty (Dierckxsens et al. Citation2017) and modified using Geneious Prime version 2020.2 (https://www.geneious.com). Then the annotation was performed by Plann (Huang and Cronk Citation2015) and Geneious Prime version 2020.2 based on the annotation of A. arguta (NC_034913.1). Finally, the validated complete chloroplast genome of A. arguta var. giraldii was deposited in Genbank (accession number MT890912).
The complete chloroplast genome of A. arguta var. giraldii is 156,729 bp in length, including a large single-copy region (LSC) of 89,647 bp and a small single-copy region (SSC) of 22,482 bp, and two 22,300 bp inverted repeat regions (IR). A total of 131 genes are annotated, containing 84 protein-coding genes, 39 tRNA genes, and 8 ribosomal RNA genes. The overall GC content of A. arguta var. giraldii chloroplast genome is 50.1%.
To confirm the phylogenetic position of A. arguta var. giraldii, ten chloroplast genome sequences of Actinidiaceae and one sequence of Saurauia were aligned by MEGA version 7.0 (Kumar et al. Citation2016). The result indicated that A. arguta var. giraldii was found to be relatively closely related to Actinidia kolomikta compared to other species of Actinidia genera in Actinidiaceae (). Our results defines the phylogenetic position of A. arguta var. giraldii at the molecular level, further improves the chloroplast genome information in Actinidia and provides fundamental information for further phylogenetic researches and exploitation, utilization of Actinidia.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT890912. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA677853, SRR13039560, and SAMN16745906, respectively.
Additional information
Funding
References
- Diana P, Cristina DM, Francisca R. 2020. Bioactivity, phytochemical profile and pro-healthy properties of Actinidia arguta: a review. Food Res Int. 136:109449.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVO Plasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1425.
- Latocha P, Łata B, Stasiak A. 2015. Phenolics, ascorbate and the antioxidant potential of kiwiberry vs. common kiwifruit: the effect of cultivar and tissue type. J Funct Foods. 19:155–163.
- Latocha P. 2017. The nutritional and health benefits of kiwiberry (Actinidia arguta) - a review. Plant Foods Hum Nutr. 72(4):325–334.
- Leontowicz H, Leontowicz M, Latocha P, Jesion I, Park YS, Katrich E, Barasch D, Nemirovski A, Gorinstein S. 2016. Bioactivity and nutritional properties of hardy kiwi fruit Actinidia arguta in comparison with Actinidia deliciosa ‘Hayward’ and Actinidia eriantha ‘Bidan’. Food Chem. 196:281–291.
- Wojdyło A, Nowicka P. 2019. Anticholinergic effects of Actinidia arguta fruits and their polyphenol content determined by liquid chromatography-photodiode array detector-quadrupole/time of flight-mass spectrometry (LC-MS-PDA-Q/TOF). Food Chem. 271:216–223.
- Zhu R, Zhang X, Wang Y, Zhang L, Zhao J, Chen G, Fan J, Jia Y, Yan F, Ning C. 2019. Characterization of polysaccharide fractions from fruit of Actinidia arguta and assessment of their antioxidant and antiglycated activities. Carbohydr Polym. 210:73–84.