Abstract
Lemmaphyllum intermedium is a valuable medicinal fern in China. We have determined its complete chloroplast genome using Illumina paired-end sequencing. The genome is 157,795 bp in length with 132 genes, including 88 protein-coding genes, eight ribosomal RNA genes, 35 tRNA genes, and one pseudogene. It has a quadripartite structure consisting of a small single-copy (SSC) of 21,764 bp, a large single-copy (LSC) of 81,189 bp, and two inverted repeats (IRs) of 27,421 bp each. A maximum-likelihood phylogenetic tree reconstructed by using complete chloroplast genome sequences reveals that L. intermedium is closely related to Lemmaphyllum carnosum var. microphyllum with strong support.
Lemmaphyllum intermedium (Ching) Li Wang, also known as Lemmaphyllum diversum (Rosenstock) Tagawa, is a vigorous fern belonging to Polypodiaceae (Zhang et al. Citation2013). The plant grows on rocks under the forest canopy at the altitude of 700 to 2200 m. Its rhizomes are covered with brown and subulate-lanceolate scales, and sporangia in discrete sori is arranged in a line on each side of midrib (Zhang et al. Citation2013). In China, L. intermedium is mainly distributed in provinces Shanxi, Gansu, Hubei, Sichuan, and Guizhou (Zhang et al. Citation2013). The whole plant of the fern is used in traditional Chinese medicine for inflammation, arthralgia, and bleeding stopping (Zhang et al. Citation2013). There exists taxonomic ambiguity for genera Lemmaphyllum, Weatherbya, Lepisorus, and Caobangia due to morphological similarity (Wang et al. Citation2010; Wei et al. Citation2017). The phylogenetic position of Lemmaphyllum and Lepidogrammitis remains in disputation (Wang et al. Citation2010). More importantly, the species delimitation in Lemmaphyllum is still an unresolved issue (Wei and Zhang Citation2013). Therefore, characterization of the whole chloroplast genome sequence of L. intermedium will be helpful to clarify the phylogeny and taxonomy of Lemmaphyllum.
Total genomic DNA was isolated from fresh and healthy fronds of L. intermedium collected from South China Botanical Garden, Chinese Academy of Sciences (23°11′3.56″N, 113°21′43.28″E). The specimen was deposited in the Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 20161024). We constructed a shotgun library with approximate insert lengths of 300 bp and performed a high-throughput sequencing on Hiseq 2500 platform (Illumina Inc., San Diego, CA). A total of 2.66 G pair-end raw data were generated. Trimmomatic v0.32 (Bolger et al. Citation2014) and FastQC v0.10.0 (Andrews Citation2010) were used for quality-trimming and assess. Then the obtained clean data were de novo assembled with Velvet v1.2.07 (Zerbino and Birney Citation2008). The assembled chloroplast genome was annotated by using GeSeq (Tillich et al. Citation2017) and tRNAscan-SE programs (Schattner et al. Citation2005), and corrected by blasting search against Lepisorus clathratus and Lemmaphyllum carnosum var. microphyllum (GenBank: KY419704 and MN623356) as references. To conduct phylogenetic analyses the complete chloroplast genome sequences of 12 ferns were retrieved from the National Center for Biotechnology Information (NCBI), and the sequence alignment was created by using the MAFFT v7.311 (Katoh and Standley Citation2013). A maximum likelihood (ML) tree was constructed with RAxML v8.2.12 (Stamatakis Citation2014) using Alsophila spinulosa as outgroup. Bootstrap values were computed from 1000 replicates.
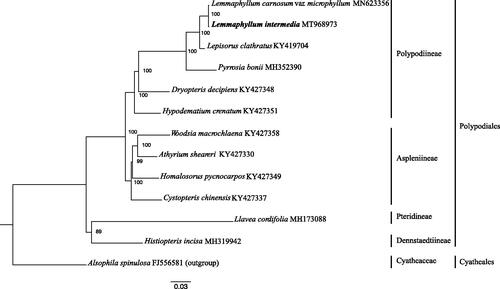
The chloroplast genome of L. intermedium is 157,795 bp in length with 132 genes (accession number: MT968973), including 88 protein-coding genes, eight ribosomal RNA genes, 35 tRNA genes, and one pseudogene. It has a typical quadripartite structure composed by a small single-copy (SSC, 21,764 bp), a large single-copy (LSC, 81,189 bp), and two inverted repeats (IRa and IRb, 27,421 bp each). Its GC content of whole genome, LSC, SSC, and IR is 42%, 41%, 38%, and 45%, respectively. There are one or two introns occurring in 18 genes: ndhB, rps16, atpF, rpoC1, ycf3 (2), clpP (2), petB, petD, rps12 (2), ndhA, rpl16, rpl2, trnG-UCC, trnV-UAC, trnA-UGC, trnI-GAU, trnL-UAA, and trnT-UGU. Fourteen genes are duplicated in the IR regions, which are rps12, rps7, psbA, ycf2, trnN-GUU, trnH-GUG, trnI-GAU, trnA-UGC, trnT-UGU, trnR-ACG, rrn4.5, rrn5, rrn16, and rrn23. ML tree reveals that L. intermedium is sister to L. carnosum var. microphyllum with a high bootstrap value (). Our chloroplast genome sequencing provides essential data for the phylogenetic characterization of Lemmaphyllum and its related genera.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MT968973, GenBank accession number MT968973. Raw sequencing reads in this study are deposited in https://www.ncbi.nlm.nih.gov/sra/SRR12989517, with SRA number SRR12989517.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data; [accessed 2020 Jul 20]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang L, Wu ZQ, Xiang QP, Heinrichs J, Schneider H, Zhang XC. 2010. A molecular phylogeny and a revised classification of tribe Lepisoreae (Polypodiaceae) based on an analysis of four plastid DNA regions. Bot J Linnean Soc. 162(1):28–38.
- Wei XP, Wei R, Zhao CF, Zhang HR, Zhang XC. 2017. Phylogenetic position of the enigmatic fern genus Weatherbya (Polypodiaceae) revisited: evidence from chloroplast and nuclear gene regions and morphological data. Int J Plant Sci. 178(6):450–464.
- Wei XP, Zhang XC. 2013. Species delimitation in the fern genus Lemmaphyllum (Polypodiaceae) based on multivariate analysis of morphological variation. J Syst Evol. 51(4):485–496.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhang XC, Lu SG, Lin YX, Qi XP, Moore S, Xing FW, Wang FG, Hovenkamp PH, Gilbert MG, Nooteboom HP, et al. 2013. Polypodiaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 758–850.