Abstract
The polyphagous cotton leafworm (Spodoptera littoralis) is one of the most destructive herbivorous insects worldwide. The present study reports the complete mitochondrial genome of S. littoralis collected from Egypt. The circular-mapping mitogenome was 15,408 bp in length with an overall A + T content of 81.1%, encoding a common set of 37 genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes. Most PCGs were found to use conventional ATN as the start codon and TAN as the stop codon. The phylogenetic tree based on the nucleic acid sequences of 13 shared PCGs of 29 Noctuidae species revealed that S. littoralis and Spodoptera litura are sister species. The data in this study will be helpful to understand geographical genetic variations, phylogenetic relationships, and species identification of S. littoralis.
The cotton leafworm, Spodoptera littoralis (Boisduval, 1833) (Lepidoptera: Noctuidae), is considered one of the most destructive herbivorous pests because of its high reproductive rate and associated heavy losses to crops. S. littoralis can feed on more than 100 host plants, potentially resulting in yield losses of 50%, which is related to its larval foliage consumption activity (El-Sheikh et al. Citation2018; Garrido-Jurado et al. Citation2020). This species is widespread in Africa, Asia, and Europe and is also classified as a quarantine pest (EPPO; Pluschkell et al. Citation1998). Strong environmental adaptability makes it a threat to countries that currently have no record of it, and monitoring is important for S. littoralis. Unfortunately, S. littoralis have similar morphology to many Spodoptera species, especially the Spodoptera litura, which making their distinction by classical physical criteria difficult (Nagoshi et al. Citation2011). Therefore, it is a great challenge for the identification and correct differentiation of S. littoralis. Mitochondrial genes such as Cytochrome c oxidase subunit 1 (cox1) are often regarded as ideal markers because of their rich distribution, maternal inheritance, and high mutation rate, and thus they are widely used in species identification, genetic diversity, and molecular phylogeny (Dormann et al. Citation2008; Bernt et al. Citation2013; Bibi et al. Citation2020; Duan et al. Citation2020). Recently, the increasing number of whole mitochondrial genomes has greatly promoted phylogenetic studies in insects (Wu et al. Citation2016; Yang et al. Citation2019b; Zhou et al. Citation2020). In our study, to accurately monitoring and understand the phylogenetic position of S. littoralis, the complete mitogenome was determined.
In this study, the genomic DNA of S. littoralis was extracted from a male moth of an inbred strain established from eggs collected near Alexandria (31°07′52.6″N 29°56′'02.5″E) in Egypt in 2011. The voucher specimen’s genomic DNA was assigned with a unique code (AGIS-SL-EG-2011) and deposited at the Agricultural Genomics Institute, Chinese Academy of Agricultural Sciences, Shenzhen, China. Total genomic DNA was extracted using the Qiagen Genomic DNA kit (Cat. no.13323, Qiagen). DNA quality and concentration were determined using NanoDrop One UV-Vis spectrophotometer (Thermo Fisher Scientific). A total of 0.5 μg of genomic DNA was used to construct a 350-bp insert library, which was then sequenced using the Illumina NovaSeq 6000 platform with 150-bp paired-end reads. The mitogenome was assembled using the software NOVOPlasty v2.5.6 (Dierckxsens et al. Citation2017). Gene annotation was performed and circularity was checked using the web-based MITOS2 (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013).
The complete mitogenome of S. littoralis (GenBank accession number MT816470) is 15,408 bp in length and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and one non-coding A + T-rich region (putative control region). In the genome, the content of A + T is 81.1% (A 41.0%; T 40.0%), which represents a typical high A + T content among insect mitogenomes (Yang et al. Citation2019a). However, the A + T content varied among genes as follows: control region (96.0%), rRNAs (84.4 ∼ 85.0%), PCGs (72.4 ∼ 92.0%), and tRNAs (70.4 ∼ 91.2%). The negligible AT-skew (0.012) and negative GC-skew (−0.198) in the mitogenome of S. littoralis were found to be similar to those of other Lepidoptera insects (Cameron and Whiting Citation2008). The total length of the 13 PCGs was determined to be 11,150 bp. All PCGs use conventional ATN as the start codon, with one exception being CGA for cox1. These special structures of the start codon are a common feature of lepidopteran mitogenomes (Wu et al. Citation2016). Ten genes are terminated with a complete stop codon (TAA), one gene (nad3) has a complete stop codon (TAG), and two genes (cox2 and nad4) end with an incomplete stop codon (T). Further, 22 tRNA genes ranged in size from 65 to 71 bp. All tRNAs harbor typical predicted secondary cloverleaf structures, except for trnN and trnY.
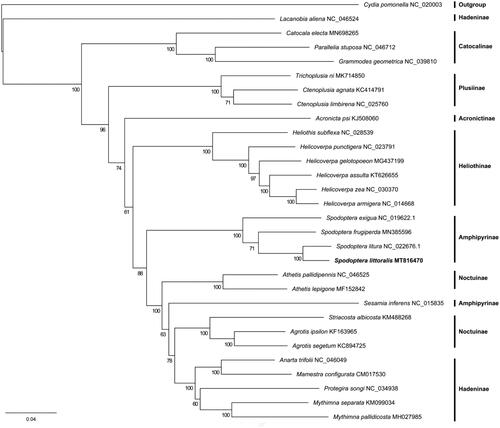
To validate the phylogenetic position of S. littoralis, we downloaded mitogenomes of 29 Noctuidae species and one outgroup species Cydia pomonella (Tortricidae) published in GenBank. Multiple nucleic acid sequence alignments of 13 PCGs were conducted using MAFFT v7.455 (Katoh and Standley Citation2013). The phylogenetic tree was constructed based on the maximum likelihood method using RAxML-NG v0.9.0 (Kozlov et al. Citation2019) with the best-fit model (GTR + F+R4) estimated by IQ-TREE v1.6.10 with parameter ‘-m MF’ (Nguyen et al. Citation2015). The bootstrap replicates were 1000 and the tree was visualized with FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). The phylogenetic analysis showed that the Spodoptera genus was monophyletic, and S. littoralis and S. litura were sister species. In summary, the current mitogenome sequence will be useful for the phylogenetic analysis of this species (). Furthermore, the mitochondrial DNA molecules within a cell are subject to evolutionary processes such as selection and drift. The high evolutionary rate of mitochondrial DNA makes it played an integral role in further revealing population structure and demographic history (Tikochinski et al. Citation2020; Johri et al. Citation2019; Hurst and Jiggins, Citation2005). S. littoralis is native to Africa and Israel (Yones et al. Citation2012), thus, the mitochondrial genome from Egypt provides the basis for further study of population genetics.
Disclosure statement
No potential conflict of interest is reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MT816470. Associated BioProject, BioSample, and SRA accession numbers are PRJNA678763, SAMN16812461, and SRR13083392, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bibi S, Khan MF, Rehman A, Shah M, Nouroz F, Nayab A. 2020. The complete mitochondrial genome of blue pansy, Junonia orithya (Lepidoptera: Nymphalidae: Nymphalinae) from Pakistan. Mitochondrial DNA B Resour. 5(3):2143–2144.
- Cameron SL, Whiting MF. 2008. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene. 408(1–2):112–123.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Dormann CF, Purschke O, García Márquez JR, Lautenbach S, Schröder B. 2008. Components of uncertainty in species distribution analysis: a case study of the Great Grey Shrike. Ecology. 89(12):3371–3386.
- Duan K, Zhang X, Zhang HH, Hu SJ. 2020. Complete mitochondrial genome of the subalpine swordtail butterfly Graphium (Pazala) parus (Nicéville, 1900) (Lepidoptera: Papilionidae). Mitochondrial DNA Part B. 5(2):1903–1904.
- El-Sheikh ESAM, El-Saleh MA, Aioub AA, Desuky WM. 2018. Toxic effects of neonicotinoid insecticides on a field strain of cotton leafworm, Spodoptera littoralis. Asian J of Biological Sciences. 11(4):179–185.
- EPPO. Spodoptera littoralis distribution and documents. EPPO Global Database; [accessed 2020 Dec 30]. https://gd.eppo.int/taxon/SPODLI.
- Garrido-Jurado I, Montes-Moreno D, Sanz-Barrionuevo P, Quesada-Moraga E. 2020. Delving into the causes and effects of entomopathogenic endophytic Metarhizium brunneum Foliar application-related mortality in Spodoptera littoralis Larvae. Insects. 11(7):e429.
- Hurst GD, Jiggins FM. 2005. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc Biol Sci. 272(1572):1525–1534.
- Johri P, Marinov GK, Doak TG, Lynch M. 2019. Population Genetics of Paramecium Mitochondrial Genomes: Recombination, Mutation Spectrum, and Efficacy of Selection. Genome Biol Evol. 11(5):1398–1416.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Nagoshi RN, Brambila J, Meagher RL. 2011. Use of DNA barcodes to identify invasive armyworm Spodoptera species in Florida. J Insect Sci. 11(1):154.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Pluschkell U, Horowitz AR, Weintraub PG, Ishaaya I. 1998. DPX-MP062- a potent compound for controlling the Egyptian cotton leafworm, Spodoptera littoralis (Boisd.). Pestic Sci. 54(2):85–90.
- Tikochinski Y, Carreras C, Tikochinski G, Vilaça ST. 2020. Author Correction: Population-specific signatures of intra-individual mitochondrial DNA heteroplasmy and their potential evolutionary advantages. Sci Rep. 10(1):13180.
- Wu YP, Zhao JL, Su TJ, Luo AR, Zhu CD. 2016. The complete mitochondrial genome of Choristoneura longicellana (Lepidoptera: Tortricidae) and phylogenetic analysis of Lepidoptera. Gene. 591(1):161–176.
- Yang M, Song L, Shi Y, Li J, Zhang Y, Song N. 2019a. The first mitochondrial genome of the family Epicopeiidae and higher-level phylogeny of Macroheterocera (Lepidoptera: Ditrysia). Int J Biol Macromol. 136:123–132.
- Yang ZH, Yang TT, Liu Y, Zhang HB, Tang BP, Liu QN, Ma YF. 2019b. The complete mitochondrial genome of Sinna extrema (Lepidoptera: Nolidae) and its implications for the phylogenetic relationships of Noctuoidea species. Int J Biol Macromol. 137:317–326.
- Yones MS, Arafat S, Abou Hadid AF, Abd Elrahman HA, Dahi HF. 2012. Determination of the best timing for control application against cotton leaf worm using remote sensing and geographical information techniques. The Egyptian Journal of Remote Sensing and Space Science. 15(2):151–160.
- Zhou Y, Zhang C, Wang SQ, Liu YL, Wang N, Liang B. 2020. A mitogenomic phylogeny of pierid butterflies and complete mitochondrial genome of the yellow tip Anthocharis scolymus (Lepidoptera: Pieridae). Mitochondrial DNA Part B. 5(3):2587–2589.