Abstract
Alniphyllum fortunei is a subtropical tree species, a large deciduous tree with a tall and straight trunk, which is an excellent fast-growing and broad-leaved tree species with a wide range of uses we resequenced complete chloroplast (cp) genome of A. fortunei from Fujian, China. The whole genome was 154,166 bp in length, consisting of a pair of inverted repeats (IR 26,658 bp), a large single-copy region (LSC 82,438 bp), and a small single-copy region (SSC 18,367 bp). The complete genome contained 139 genes, including 89 protein-coding genes, 40 tRNA, and 8 rRNA genes. The phylogenetic analyses based on the complete chloroplast genome sequence provided solid evidence that A. fortunei has a close relationship with A. pterospermum and Bruinsmia polysperma.
Alniphyllum fortunei (Hemsl.) Makino are the main timber species in southern China. A. fortunei is mainly distributed in various provinces in the south of China’s Yellow River Basin. The current main producing area is Jiangxi, Zhejiang, and Fujian Province (Hu et al. Citation2001; Wang et al. Citation2018). However, for such an important species, most of the studies focused on the cultivation and distribution pattern. Moreover, although there were some studies on molecular biology, there was nearly no report of complete chloroplast genome of Alniphyllum. Here, we reported the complete chloroplast genome sequences of A. fortunei, so as to reveal phylogenetic relationships between the species and related group in Styracaceae.
Leaf sample of A. fortunei was collected from Jian Ou, Fujian Province, China (27°16′07″N, 118°33′59″E), and dried into silica gel immediately. The voucher specimen is kept at the Herbarium of College of Forestry, Fujian Agriculture and Forestry University (Wen-Jun Lin and [email protected]) under the voucher number Afo_Linwj. Total genomic DNA were extracted from fresh leaves by using the improved CTAB method (Doyle Citation1987). The obtained DNA was fragmented to construct a paired-end library with an insert-size of 500 bp and the genome sequencing were performed using Illumina Hiseq Xten platform, approximately 5GB data generated. Illumina data were filtered by script in the cluster (default parameter: -L 5, -p 0.5, -N 0.1). Chloroplast genome of A. fortunei was assembled by Get Organelle pipe-line (https://github.com/Kinggerm/GetOrganelle), it can get the plastid-like reads, and the reads were viewed and edited by Bandage (Wick et al. Citation2015). Assembled chloroplast genome annotation base on comparison with C. echinocarpa by Geneious v 11.1.5 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012). The annotation result was drawn with the online tool OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013).
The complete chloroplast genome sequence of A. fortunei (GeneBank accession: MW042695) was 154,166 bp in length, with a large single-copy (LSC) region of 82,438 bp, a small single-copy (SSC) region of 18,367 bp, and a pair of inverted repeats (IR) regions of 26,658 bp. Complete chloroplast genome contain 139 genes, there were 89 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. The complete genome GC content was 37.1%.
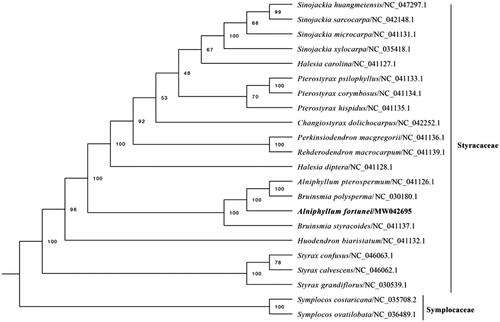
To investigate the phylogenetic position of A. fortunei, 22 complete cp genomes of species from Styracaceae and Symplocaceae, downloaded from NCBI, were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). A maximum-likelihood (ML) tree was constructed on XSEDE (8.2.12) (https://www.phylo.org/portal2/login!input.action) with 1000 bootstrap replicates. RAxML-HPC2 (Stamatakis Citation2014) was used to construct a maximum-likelihood tree with Symplocos costaricana and Symplocos ovatilobata as outgroup ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are available in [NCBI] at [https://www.ncbi.nlm.nih.gov/], Reference no. MW042695.
Additional information
Funding
References
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Hu SZ, Jiang YF, Deng GH, Yu LG,Lei XG. 2 001. Cultivation techniques of Alniphyllum fortunei[J]. Jiangxi Forestry Sci Technol. 000(006):7–9.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang L-L, Zhang Y, Yang Y-C, Du X-M, Ren X-L, Liu W-Z. 2018. The complete chloroplast genome of Sinojackia xylocarpa (Ericales: Styracaceae), an endangered plant species endemic to China. Conserv Genet Resour. 10(1):51–54.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.