Abstract
Lonicera hypoglauca Miq, which is widely distribute in south China, is an important Chinese plant used in traditional medicine. Here we report the first complete chloroplast (cp) genome sequence of this species. The circular cp genome is 154,581 bp in size, including a large single-copy (LSC) region of 88,379 bp and a small single-copy (SSC) region of 18,646 bp, which were separated by two inverted repeat (IR) regions (IRA and B, 23,778 bp each). A total of 121 genes were annotated, including 8 ribosomal RNAs (rRNAs), 33 transfer RNAs (tRNAs) and 80 protein-coding genes (PCGs). Phylogenetic analysis of 20 representative members within the Caprifoliaceae showed that L. hypoglauca is closely related to the Lonicera macranthoides. This study provides important genetic information for future systematic and evolutionary studies of L. hypoglauca.
Lonicera hypoglauca Miq, which is classified in the Caprifoliaceae, is widely distributed in southern China and Japan. The flower bud (rich in chlorogenic acid) of this plant is used as an herbal medicine and has been prescribed to treat various infectious diseases, such as fever, febrile blood dysentery and throat numbness (Zhang et al. Citation2008; Li et al. Citation2015). Pheophytin a, which can be extracted from the L. hypoglauca leaves, is an effective inhibitor of chronic hepatitis C virus NS3 protease (Wang et al. Citation2009). Some compounds (Bisflavonoid and loniceraflavone) in the leaves of L. hypoglauca significantly inhibit the activity of xanthine oxidase (Chien et al. Citation2009). Although the medicinal effect of L. hypoglauca have been fully studied, the complete chloroplast (cp) genome of L. hypoglauca has not been reported. In this research, we assembled and determined the cp genome sequence of L. hypoglauca as a resource for future study of this medicinal plant.
Young and healthy leaf samples were collected from Guangxi Botanical Garden of Medicinal Plants, Nanning, Guangxi Zhuang Autonomous Region, China (22°51′17.77″N, 108°24′48.15″E, 95 m above sea level). The leaf specimen (accession number: GZNUYZW202002001) was deposited in the herbarium of School of Life Sciences, Guizhou Normal University (Jinghua Mu, [email protected]). The total genomic DNA (No. YZW202002002) was extracted using the DNAsecure Plant Kit (TIANGEN, Beijing) and stored at −80 °C in the laboratory (room number: 1403) of School of Life Science, Guizhou Normal University. A total concentration of 800 ng of DNA served as the input material for the DNA sample preparations. Sequencing libraries were generated using NEB Next® Ultra DNA Library Prep Kit for Illumina® (NEB, USA). The library preparations were sequenced on an Illumina platform and 150 bp paired-end reads were generated. The filtered reads were assembled using the program GetOrganelle (Jin et al. Citation2019) with Lonicera macranthoides (GenBank accession number: MH579750) as the reference genome. The assembled cp genome was annotated using the online software GeSeq (Tillich et al. Citation2017). The complete chloroplast genome sequence was submitted to GenBank under the accession number MW186761.
The length of the complete cp genome sequence of L. hypoglauca is 154,581 bp, consisting of a large single-copy (LSC, 88,379 bp) region, a small single-copy (SSC, 18,646 bp) region and two inverted repeats (IRA and IRB) regions of 23,778 bp each. In total, 121 genes were predicted, including 80 protein coding (PCGs), 8 rRNA and 33 tRNA genes. Among the assembled genes, 4 rRNAs (rrn16, rrn23, rrn4.5 and rrn5), 2 PCGs (rps7 and ndhB) and 7 tRNAs (trnA-UGC, trnH-GUG, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC) occurred as double copies. One PCG (rps12) occurred in three copies. Intron-exon analysis showed the majority (102 genes, 84%) of the genes do not display introns, whereas 19 (16%) genes contained introns.
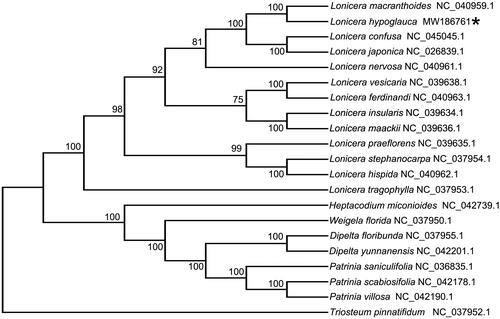
To further understand the phylogenetic history of L. hypoglauca, 20 cp genome sequences of the Caprifoliaceae (12 Lonicera species, 1 Heptacodium species, 1 Triosteum specie, 2 Dipelta species, 1 Weigela species and 3 Patrinia species) were downloaded from GenBank to construct the phylogenetic trees through maximum-likelihood (ML) analysis. The ML tree was performed using RAxML (Version 8.0.19, Model: GTRGAMMA) (Stamatakis Citation2014) with 1,000 bootstrap replicates. The phylogenetic tree indicated that L. hypoglauca was fully resolved in a clade with L. macranthoides, sister to two other species of Lonicera, L. confusa and L. japonica (). Compared to other Flos Lonicerae members, L. hypoglauca was also fully resolved in a clade with L. macranthoides according to rbcL gene sequence analysis (Li et al. Citation2012). Because of the closely evolutionary relationship between L. macranthoides and L. hypoglauca, in comparison with other DNA barcodes, only the psbA-trnH intergenic spacer sequence had appropriate mutation sites to distinguish L. macranthoides and L. hypoglauca (Sun et al. Citation2011).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession number MW186761. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA674956, SRX9460983, and SAMN16684231, respectively.
Additional information
Funding
References
- Chien SC, Yang CW, Tseng YH, Tsay HS, Kuo YH, Wang SY. 2009. Lonicera hypoglauca inhibits xanthine oxidase and reduces serum uric acid in mice. Planta Med. 75(4):302–306.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. 256479.
- Li X, Tao Z, Wu Z, Lin X, Fan C. 2012. RbcL sequence analysis of Lonicera in the South of Zhejiang province. J Wenzhou Med College. 42:549–552.
- Li Y, Cai W, Weng X, Li Q, Wang Y, Chen Y, Zhang W, Yang Q, Guo Y, Zhu X, et al. 2015. Lonicerae Japonicae Flos and Lonicerae Flos: a systematic pharmacology review. Evid Based Complement Alternat Med. 2015:905063.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun Z, Gao T, Yao H, Shi L, Zhu Y, Chen S. 2011. Identification of Lonicera japonica and its related species using the DNA barcoding method. Planta Med. 77(3):301–306.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang SY, Tseng CP, Tsai KC, Lin CF, Wen CY, Tsay HS, Sakamoto N, Tseng CH, Cheng JC. 2009. Bioactivity-guided screening identifies pheophytin a as a potent anti-hepatitis C virus compound from Lonicera hypoglauca Miq. Biochem Biophys Res Commun. 385(2):230–235.
- Zhang B, Yang R, Zhao Y, Liu CZ. 2008. Separation of chlorogenic acid from honeysuckle crude extracts by macroporous resins. J Chromatogr B Analyt Technol Biomed Life Sci. 867(2):253–258.