Abstract
The complete chloroplast (cp) genome of Procris crenata C.B.Rob was reported. The cp genome was 154,124 bp in length and contained two inverted repeats (IRs) of 25,626 bp, which were separated by large single-copy and small single-copy of 84,599 bp and 18,273 bp, respectively. The GC content was 36.5%. A total of 113 functional genes were encoded, comprising 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. This is the first reported plastid genome in Procris (Urticaceae), which will be useful data for resolving the relationship within the family.
Keywords:
Procris Comm. ex Juss. (Urticaceae) is a small genus with about 20 species of subshrubs, shrubs, epiphytic, and epilithic (Chen et al. Citation2003). It is distributed throughout warm-temperate and tropical regions of the Old World (Chen et al. Citation2003). As the member of tribe Elatostemteae, Procris has long been controversial with respect to Elatostema J.R.Forst. & G.Forst., Elatostematoides C.B.Rob., and Pellionia Gaudich (Hadiah et al. Citation2003, Citation2008; Hadiah and Conn Citation2009; Wu et al. Citation2013). Using three DNA regions, Tseng et al. (Citation2019) conducted an extensive phylogenetic study demonstrating that Procris is a monophyletic genus and also clarifying the genus level of Procris, Elatostema, and Elatostematoides. However, the phylogenetic relationships within Procirs still remained poorly resolved which hampered our understanding on the evolution of the genus and the family.
Plastome provides sufficient information to resolve the phylogenetic relationships at generic and infrageneric level. Despite several plastid genomes of species in Urticaceae have been reported (Wu et al. Citation2017; Wu et al. Citation2018; Fu et al. Citation2019; Wang et al. Citation2020), none of them belong to Procris.
In this study, leaves of Procris crenata C.B.Rob were collected from Longzhou County, Guangxi, China (N106°46′1″, E22°44′24″). A specimen was deposited at the herbarium of Guangxi Institute of Botany (IBK, http://www.gxib.cn/spIBK/, contact person and email: Long-Fei Fu, [email protected]) under the voucher number XZB20180127-10. Genomic DNA was extracted using the modified CTAB method (Chen et al. Citation2014) and sent to Majorbio Company (http://www.majorbio.com/, China) for next-generation sequencing. Short-insert (350 bp) paired-end read libraries preparation and 2 × 150 bp sequencing were performed on an Illumina (HiSeq4000) genome analyzer platform. Raw data (approximately 2 Gb) was filtered using the FASTX-Toolkit to obtain high-quality clean data (http://hannonlab.cshl.edu/fastx_toolkit/download.html). The original data (SRR12846062) were mapped to plastid genome reference (Elatostema dissectum, GenBank-MF227819) in Geneious Primer (Kearse et al. Citation2012) to exclude nuclear and mitochondrial reads. Putative plastid reads were then used for de novo assembling construction. Contigs were able to be concatenated by the overlaps using the Repeat Finder program implemented in Geneious Primer until a ∼130kb contig (including SSC, IR and LSC) being built. The IR region was determined by the Repeat Finder program in Geneious Primer and was reverse copied to obtain the complete chloroplast genome. The annotation approach was performed using CPGAVAS2 and PGA (Qu et al. Citation2019; Shi et al. Citation2019).
The complete chloroplast genome of Procris crenata was 154,124 bp in length (GenBank-MW114890), the GC content was 36.5%. Large single-copy and small single-copy were 84,599 bp and 18,273 bp respectively, while IR was 25,626 bp in length. The plastid genome encoded 113 functional genes, including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes.
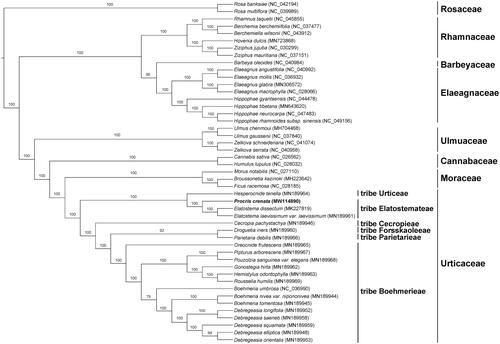
The maximum likelihood phylogeny was reconstructed by the dataset of 80 encoded protein genes from 21 species of Urticaceae and 26 species of other families in Rosales (). The result was congruent with previous studies showing that Procris was closely related to Elatostema. And they together formed a clade representing tribe Elatostemateae which was sister to tribe Urticeae (Wu et al. Citation2013; Tseng et al. Citation2019). The newly reported plastid genome will provide additional data for further study on the phylogeny and evolution of the genus and the family.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW114890. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA669252, SRS7538264, and SAMN16443195 respectively.
Additional information
Funding
References
- Chen CJ, Friis I, Wilmot-Dear CM. 2003. Procris. In: Wu ZY, Raven PH, editors. Flora of China 5. Beijing: Science Press & Missouri Botanical Garden Press; p. 163.
- Chen LY, Song MS, Zha HG, Li ZM. 2014. A modified protocol for plant genome DNA extraction. Plant Divers Resour. 36:375–380.
- Fu LF, Xin ZB, Wen F, Li S, Wei YG. 2019. Complete chloroplast genome sequence of Elatostema dissectum (Urticaceae). Mitochondrial DNA Part B. 4(1):838–839.
- Hadiah JT, Conn BJ, Quinn CJ. 2008. Infra-familial phylogeny of Urticaceae, using chloroplast sequence data. Aust Systematic Bot. 21(5):375–385.
- Hadiah JT, Conn BJ. 2009. Usefulness of morphological characters for infrageneric classification of Elatostema (Urticaceae). Blum – J Plant Tax Plant Geog. 54(1):181–191.
- Hadiah JT, Quinn CJ, Conn BJ. 2003. Phylogeny of Elatostema (Urticaceae) using chloroplast DNA data. Telopea. 10(1):235–246.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Qu X, Moore MJ, Li D, Yi T. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Tseng YH, Monro AK, Wei YG, Hu JM. 2019. Molecular phylogeny and morphology of Elatostema s.l. (Urticaceae): implications for inter- and infrageneric classifications. Mol Phylogenet Evol. 132:251–264.
- Wang RN, Milne RI, Du XY, Liu J, Wu ZY. 2020. Characteristics and mutational hotspots of plastomes in Debregeasia (Urticaceae). Front Genet. 11:729.
- Wu ZY, Du XY, Milne RI, Liu J, Li DZ. 2017. Charaterization of the complete chloroplast genome sequence of Cecropia pachystachya. Mitochondrial DNA Part B. 2(2):735–737.
- Wu ZY, Du XY, Milne RI, Liu J, Li DZ. 2018. Complete chloroplast genome sequences of two Boehmeria species Urticaceae. Mitochondrial DNA Part B. 3:939–940.
- Wu ZY, Monro AK, Milne RI, Wang H, Yi TS, Liu J, Li DZ. 2013. Molecular phylogeny of the nettle family (Urticaceae) inferred from multiple loci of three genomes and extensive generic sampling. Mol Phylogenet Evol. 69(3):814–827.