Abstract
Ephedra monosperma is an important medicinal plant of Ephedra (Ephedraceae). The complete chloroplast genome of E. monosperma was assembled from Illumina pair-end sequence reads. The whole chloroplast (cp) genome is 109,548 bp in length and presents a quadripartite structure consisting of two copies of inverted repeat (IR) regions (20,398) separated by a large single copy (LSC) region (60,674 bp) and a small single copy (SSC) region (8078 bp). The cp genome of E. monosperma encodes a total of 118 genes, including 73 protein-coding genes, 37 tRNA genes and 8 rRNA genes. The overall GC content of E. monosperma cp genome is 36.6%. A maximum likelihood (ML) phylogenetic analysis revealed that E. monosperma was close to Ephedra equisetina. The ML tree also showed Ephedraceae appeared more closely related to Gnetaceae than to the other families in Gymnospermae.
Ephedra plants have been used as medicines for several thousand years (Editorial Committee of Zhonghua Bencao National Traditional Chinese Herb Administration Citation1996; Caveney et al. Citation2001). Ephedra monosperma Gmel. ex C.A. Mey., a drought-tolerant shrub, has high content of ephedrine and pseudoephedrine (Ibragic and Sofić Citation2015). To date, there have been few studies for this medicinally and ecologically important plant (e.g. Sofronova et al. Citation2014;Ibragic and Sofić Citation2015). In this study, we report and characterize the complete chloroplast (cp) genome of E. monosperma (GenBank accession number: MW186779) to provide genomic data for future research.
An individual of E. monosperma was collected in Chunkun Mountain, Baotou, Inner Mongolia, China (E110°44′, N41°17′). The specimen was deposited at the herbarium of Baotou Teachers’ College (email: [email protected]) under the voucher number CKS-2020-07-B07DZMH. Total genomic DNA was isolated from silica-dried leaf material using modified CTAB method (Doyle Citation1987), and then sequenced using an Illumina Hiseq 2500 platform at Biomarker Technologies Inc. (Beijing, China). A total of 574,210 reads were assembled to generate the cp genome of E. monosperma. Reference-guided assembly was used to reconstruct the cp genomes with the programs MIRA 4.0.2 (Chevreux et al. Citation2004) and MITObim v1.7 (Hahn et al. Citation2013). In the process, cp genome of Ephedra intermedia (NC_044772) was used as reference genome. The complete cp genome was annotated in GENEIOUS R10 (Biomatters Ltd., Auckland, New Zealand), and then manually corrected by comparing with the published complete cp genomes (GenBank accession numbers: NC_011954, NC_044772) of Ephedra species using GENEIOUS R10.
The complete cp genome of E. monosperma is 109,548 bp in length with a quadripartite structure. The large single copy (LSC) region, small single copy (SSC) region and inverted repeat (IRa and IRb) regions are 60,674 bp, 8078 bp and 20,398 bp, respectively. The assembled cp genome encodes a total of 118 genes, consisting of 73 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of E. monosperma cp genome is 36.6%, and the corresponding values in LSC, SSC and IR regions are 34.2%, 27.4%, and 42.1%, respectively.
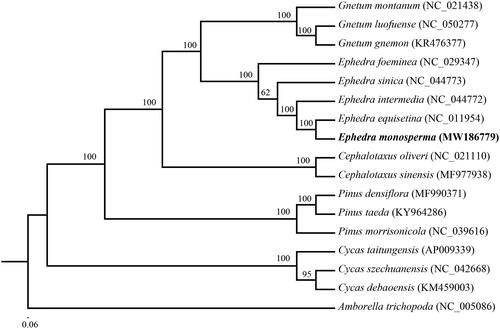
The phylogenetic tree was constructed based on 17 complete cp genomes of vascular plants. All of the 17 complete cp genome sequences were aligned using MAFFT (Katoh and Standley Citation2013) with default parameter. To reconstruct the phylogenetic relations of these species we used the GTR + G model with maximum likelihood (ML) analysis. A ML tree was constructed with RAxML v8 (Stamatakis Citation2014). The node stability was assessed using a rapid bootstrapping analysis with 1000 replicates. The phylogenetic tree indicated that E. monosperma was close to Ephedra equisetina with high support (). The ML tree also showed Ephedraceae appeared more closely related to Gnetaceae than to the other families in Gymnospermae, which is consistent with previous molecular results (Chen et al. Citation2019). The cp genome of E. monosperma will provide useful genetic information for further study on genetic diversity and conservation of Ephedraceae species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW186779. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA681937, SRP295568, and SAMN16975003, SAMN16975004, respectively.
Additional information
Funding
References
- Caveney S, Charlet DA, Freitag H, Maier‐Stolte M, Starratt AN. 2001. New observations on the secondary chemistry of world Ephedra (Ephedraceae). Am J Bot. 88(7):1199–1208.
- Chen XL, Cui YX, Nie LP, Hu HY, Xu ZC, Sun W, Gao T, Song JY, Yao H. 2019. identification and phylogenetic analysis of the complete chloroplast genomes of three Ephedra herbs containing ephedrine. Biomed Res Int. 2019:5921725.
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14(6):1147–1159.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Editorial Committee of Zhonghua Bencao National Traditional Chinese Herb Administration. 1996. Zhonghua Bencao (Chinese Herbal Medicine). Vol. 2. Shanghai: Shanghai Science &Technology Press.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucl Acids Res. 41(13):e129–e129.
- Ibragic S, Sofić E. 2015. Chemical composition of various Ephedra species. Bosn J Basic Med Sci. 15(3):21–27.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Sofronova VE, Chepalov VA, Dymova OV, Golovko TK. 2014. The role of pigment system of an evergreen dwarf shrub Ephedra monosperma in adaptation to the climate of Central Yakutia. Russ J Plant Physiol. 61(2):246–254.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.