Abstract
The nearly complete mitochondrial genome (mitogenome) of Xenos cf. moutoni, one twisted-wing parasite on wasp Vespa velutina from Southwest of China, is described in this study. The total length of this mitogenome is 16,717 bp, containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs) and an incomplete A + T-rich control region . All of the 13 PCGs are initiated with canonical ATN (N represents A, T, G, C) as start codons; 8 PCGs are terminated with a complete typical stop codon TAA, and the remaining five PCGs (cox2, cox3, nad3, nad4 and nad5) have an incomplete stop codon with just a T. The phylogenetic analysis based on the nucleotide sequences of PCGs and rRNAs indicates that Xenos cf. moutoni has a close relationship with Xenos vesparum, confirming its placement in the family Xenidae.
Strepsiptera, often known as ‘twisted wing’ parasitoids, is a kind of enigmatic insects with a complex life cycle and unusual morphology (Pohl and Beutel Citation2008; Cook Citation2014; Kathirithamby Citation2018). It is a small order of holometabolous insects, comprising 630 species in 15 families, five of which are extinct (Kathirithamby Citation2018). All strepsipterans utilize insects as hosts, and their host associations are found in seven insect orders (Cook Citation2014). Among 10 extant families, the family Mengenillidae use apterygote insect (Zygentoma) as host, and the families, Stylopidae and Xenidae, are known to use aculeate Hymenoptera as hosts: Stylopidae parasitizes bees (Apidae), and Xenidae parasitizes wasps (Crabronidae, Sphecidae, and Vespidae) (Pohl and Beutel Citation2008; Cook Citation2014). The Xenidae includes ca. 110 described species (ca. 19%) and thus is one of the species-rich strepsipteran families (Pohl and Beutel Citation2008). To date, only three mitogenomic sequences (Mengenilla australiensis and M. moldrzyki in Mengenillidae and Xenos vesparum in Xenidae) were reported (Carapelli et al. Citation2006; McMahon et al. Citation2009; Niehuis et al. Citation2012).
Twisted-wing insect species Xenos cf. moutoni (males and famales) were collected from the body of wasps (Hymenoptera, Vespidae, Vespa velutina) in one nest was collected from Gaoligong Mountains, Xiangda Township, Longling County, Yunnan Province (98°43′26″E, 24°26′38″N), China, on 20 December 2019 by local villagers. With reference to male morphological traits of strepsipteran insects (Buysson Citation1903; Kifune and Maeta Citation1985; Kathirithamby and Engel Citation2014) and the published strepsipteran mitochondrial cox1 data (Benda et al. Citation2019), this species is identified as Xenos cf. moutoni in the family Xenidae. The voucher specimens (voucher no. KIZ0127514) are stored in Kunming Natural History Museum of Zoology, Chinese Academy of Sciences. Total genomic DNA (gDNA) of one female adult was isolated using a Gentra Puregene Blood kit (Qiagen, Hilden, Germany) based on instructions. Library (150-bp insert size) was prepared and sequenced on Illumina Novaseq 6000 (Novogene, Beijing, China). The 126,444,619 clean reads obtained were used to execute the assembly using both MitoZ v2.3 (Meng et al. Citation2019) and NOVOPlasty v4.2 (Dierckxsens et al. Citation2017). Gene annotation was performed using the MITOS2 webserver (Bernt et al. Citation2013) with manual correction.
Both assembled methods (MitoZ and NOVOPlasty) produced the same sequence except A + T-rich region which is longer in NOVOPlasty assembly than in MitoZ assembly. The mitogenome of Xenos cf. moutoni (GenBank accession No. MW222190) using NOVOPlasty was 16,717 bp in length and 1,658,750 reads were mapped to the mitogenome with 6,904 mean coverage. The linear mitogenome was considered to be incomplete because its two terminuses lack repetitive overlaps with longer than 25 bp (Tang et al. Citation2014). It contained 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs) and an incomplete control (A + T-rich) region (about 2107 bp). All 13 PCGs are 10,676 bp in length which contain 35.59% A, 9.49% G, 44.30% T and 10.62% C with a strong bias toward A + T (79.89%). All of 13 PCGs use ‘ATN’ (N represents A, T, G, C) as the start codons; eight of them are terminated with typical stop codon ‘TAA’, and the other five (cox2, cox3, nad3, nad4 and nad5) have an incomplete stop codon ‘T––’. The length of 22 tRNAs ranges from 56 bp to 69 bp and the size of small rRNA (s-rRNA) and large rRNA (l-rRNA) is 735 bp and 1234 bp, respectively. The arrangement of all 13 PCGs in Xenos cf. moutoni is consistent with those of two other strepsipterans (M. australiensis and X. vesparum) and the luminous beetle R. lufengensis (Carapelli et al. Citation2006; Li et al. Citation2007; McMahon et al. Citation2009). However, some tRNA gene order changes are observed among the four species in those regions between nad3 and nad5, nad5 and nad4 as well as s-rRNA and nad2 and in the location of the trnL1 (Carapelli et al. Citation2006; Li et al. Citation2007; McMahon et al. Citation2009).
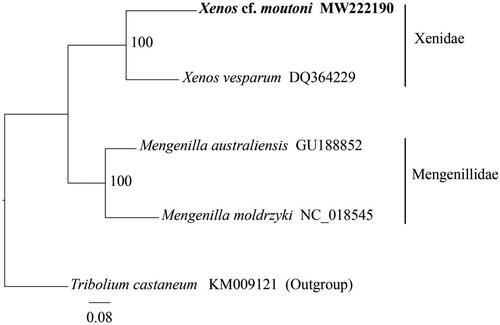
Phylogenetic interference was performed among Xenos cf. moutoni and other three reported strepsiperan species (Carapelli et al. Citation2006; McMahon et al. Citation2009; Niehuis et al. Citation2012) with Tribolium castaneum (Coleopetera, Tenebrionidae) as an outgroup. The 13 PCGs and two rRNAs from these species were aligned and concatenated using MEGA-X (Kumar et al. Citation2018) and then trimmed the unreliably regions using trimAl v1.4 (gt = 0.5) (Capella-Gutierrez and Silla-Martinez Citation2009). The phylogenetic tree was constructed based on the concatenated nucleotide sequence using RAxML v8.2.10 (Stamatakis Citation2014) with the GTR + G + I model. Our result indicates that Xenos cf. moutoni has a close relationship with X. vesparum in the family Xenidae with high bootstrap values (), confirming its placement in the family Xenidae.
Figure 1. The phylogenetic tree of four strepsipteran species based on the nucleotide sequence of concatenated 13 protein-coding genes (PCGs) and two ribosomal RNA genes (rRNAs) using maximum likelihood (ML) analysis. Red flour beetle Tribolium castaneum was selected as the outgroup. The values at the nodes show the bootstrap support calculated using the maximum likelihood method with 100 replicates.
Acknowledgements
The authors thank Dr. Jiangli Tan for helping in wasp identification and Drs Ping Hu, Delong Guan, Jie Yang and Cheng Jin for discussion in data analysis. We also thank those anonymous villagers for helping in collecting parasited wasps. LXY and WW designed this study. ZR, LXY and LJ wrote this manuscript. ZR, MCY and HJW made data analysis. DZW and LXY collected and identified specimens. LGC and ZRP carried out experiments.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, under the accession no. MW222190. The associated BioProject, Bio-Sample and SRA numbers are PRJNA681068, SAMN16898475 and SRR13154212, respectively.
Additional information
Funding
References
- Benda D, Nakase Y, Straka J. 2019. Frozen Antarctic path for dispersal initiated parallel host-parasite evolution on different continents. Mol Phylogenet Evol. 135:67–77.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Buysson RD. 1903. Note pour servir à l’histoire des Strepsiptères. Bull Soc Entomol France. 8(9):174–175.
- Capella-Gutierrez S, Silla-Martinez JMT. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Carapelli A, L, Vannini, F, Nardi, JL, Boore, L, Beani, R, Dallai, F. Frati 2006. The mitochondrial genome of the entomophagous endoparasite Xenos vesparum (Insecta: Strepsiptera). Gene. 376(2):248–259.
- Cook JL. 2014. Review of the biology of parasitic insects in the order Strepsiptera. Compar Parasitol. 81(2):134–151.
- Dierckxsens N, P, Mardulyn, G. Smits 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18
- Kathirithamby J. 2018. Biodiversity of Strepsiptera. In: Foottit RG, Adler PH, editors. Insect biodiversity: science and society. Vol II. New York: John Wiley & Sons; p. 673–703.
- Kathirithamby J, MS. Engel 2014. A revised key to the living and fossil families of Strepsiptera, with the description of a new family, Cretostylopidae. J Kansas Entomol Soc. 87(4):385–388.
- Kifune T, Maeta Y. 1985. Taxonomical studies on the genus Xenos (Strepsiptera, Stylopidae) parasitic on vespa and polistes (Hymenoptera, Vespidae) of Taiwan with descriptions of three new species. Japanese J Entomol. 53(3):426–435.
- Kumar S, G, Stecher, M, Li, C, Knyaz K. Tamura 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li X, K, Ogoh, N, Ohba. X, Liang, Y. Ohmiya 2007. Mitochondrial genomes of two luminous beetles, Rhagophthalmus lufengensis and R. ohbai (Arthropoda, Insecta, Coleoptera). Gene. 392(1–2):196–205.
- McMahon DP, A, Hayward, J. Kathirithamby 2009. The mitochondrial genome of the 'twisted-wing parasite' Mengenilla australiensis (Insecta, Strepsiptera): a comparative study. BMC Genom. 10(1):603
- Meng GL, YY, Li, CT, Yang, SL. Liu 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Niehuis O, G, Hartig, S, Grath, H, Pohl, J, Lehmann, H, Tafer, A, Donath, V, Krauss, C, Eisenhardt, J, Hertel, et al. 2012. Genomic and morphological evidence converge to resolve the enigma of Strepsiptera. Curr Biol. 22(14):1309–1313.
- Pohl H, RG. Beutel 2008. The evolution of Strepsiptera (Hexapoda). Zoology. 111(4):318–338.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tang M, M, Tan, G, Meng, S, Yang, X, Su, S, Liu, W, Song, Y, Li, Q, Wu, A, Zhang, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes – a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42(22):e166.
