Abstract
Nephelium lappaceum is a popular tropical fruit belonging to the Sapindaceae family. The plant originated in Malaysia and Indonesia and is commonly called rambutan. Because of its refreshing flavor and exotic appearance, rambutan is widely accepted in the World. Due to its significant medicinal properties, the fruit has also been employed in traditional medicine for centuries. The chloroplast genome of rambutan was sequenced, assembled, and annotated in the present study. The chloroplast genome length was 161,356 bp and contained 132 genes, including 87 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. It possessed the typical quadripartite circle structure with a large single-copy region (86,009 bp), a small single-copy region (18,153 bp), and two inverted repeat regions (28,597 bp). A total of 35 SSR markers were found in the chloroplast genome of Nephelium lappaceum, of which 33 were monomer, 1 was dimer and 1 was tetramer. Phylogenetic analysis based on the complete chloroplast genome sequences of 21 plant species showed that rambutan was closely related to Pometia tomentosa. These results provide a foundation for further phylogenetic and evolutionary studies of the Sapindaceae family.
Nephelium lappaceum Linn. 1767, commonly called rambutan, is a popular tropical fruit of the Sapindaceae family that originated in Malaysia (Wall Citation2006; Odey Citation2012). In terms of phylogenetic relationship, rambutan is closely related to lychee, pulasan, and longan (Greenand Popenoe Citation1975). Although rambutan is a fruit of Asian origin, it is widely appreciated in other parts of the World due to its refreshing flavor and exotic appearance, where it is consumed fresh, canned, or processed (Almeyada et al. Citation1979). Rambutan also has medicinal properties (Soeng et al. Citation2015; Mahmood et al. Citation2018); the rind extract exhibits high anti-oxidant (Palanisamy et al. Citation2008; Nurhuda et al. Citation2013), antibacterial (Thitilertdecha et al. Citation2010; Yuvakkumar et al. Citation2014), anti-Herpes Simplex virus type 1 (Nawawi et al. Citation1999) and anti-hyperglycemic (Palanisamy et al. Citation2011) activities. In Southeast Asia, the dried fruit rind has been employed in traditional medicine for centuries (Phang et al. Citation2019).
Despite its significance, genomics research on rambutan is very little. The chloroplast genome contains a large amount of genetic information related to various biological processes such as photosynthesis, which is crucial in studying plant evolution, genetic relationships, and accurate evaluation of germplasm resources (Niu et al. Citation2020). This study is the first report on the sequencing, assembly, and annotation of the rambutan chloroplast genome.
Young leaves of rambutan were collected from the Xishuangbanna Tropical Flowers and Plants Garden (100.786425 E, 22.011135 N), Yunnan province of China, and the specimen was deposited at the herbarium of the Yunnan Institute of Tropical Crops (http://www.yitc.com.cn/, [email protected]) under the voucher number of YITC-2020-FZ-S-006. The genomic DNA was extracted by a DNeasy Plant Mini Kit (Qiagen. Hilden, Germany), and the DNA quality was evaluated by a Nano-Drop 2000 spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). DNA library for sequencing was constructed with insert sizes of 350 bp, and paired-end (PE) sequencing was conducted on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA). Subsequently, approximately 7.5 Gb of raw data were obtained and then assembled by GetOrganelle (Jin et al. Citation2020). The chloroplast genome was annotated by GeSeq (Tillich et al. Citation2017), examined manually by Geneious 11.1.5 (Kearse et al. Citation2012), and submitted to the GenBank under accession number MT884002.
The length of the rambutan chloroplast genome was 161,356 bp with a 37.78% GC content, and the genome structure was relatively comparable to that of other plant species (Liu et al. Citation2018). It possessed the typical quadripartite circle structure with two identical copies of inverted repeat regions (IRs, 28,597 bp) separated by a large single-copy region (LSC, 86,009 bp) and a small single-copy region (SSC, 18,153 bp). The GC contents of the LSC, IRs, and SSC regions were 36.03%, 42.28%, and 31.86%, respectively. The circular genome contained 132 genes, including 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The protein-coding genes are involved in photosystem I, photosystem II, the cytochrome b/f complex, ATP synthase, NADH dehydrogenase, RNA polymerase, and other biological functions. A total of 11 protein-coding genes, including rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, and ndhA, contained one intron, while four protein-coding genes, including rps12, ycf3, and clpP, contained two introns. The five smallest protein-coding genes were petN, petL, psbM, psbT, and psbI, and their sizes were 90 bp, 96 bp, 105 bp, 108 bp, and 111 bp. Meanwhile, the five largest protein-coding genes were ycf2, ycf1, rpoC2, rpoB, and psaA, and their sizes were 6855 bp, 5712 bp, 4182 bp, 3213 bp, and 2253 bp, respectively.
The MISA software (https://webblast.ipk-gatersleben.de/misa/) was used to analyze the microsatellite SSR, during the analysis, the minimum numbers of repeats were set to 10, 6, 5, 5, 5, and 5 for monomer, dimer, trimer, tetramer, pentamer, and hexamer, respectively. A total of 35 SSR markers were found in the chloroplast genome of Nephelium lappaceum, of which 33 were monomer, 1 was dimer and 1 was tetramer.
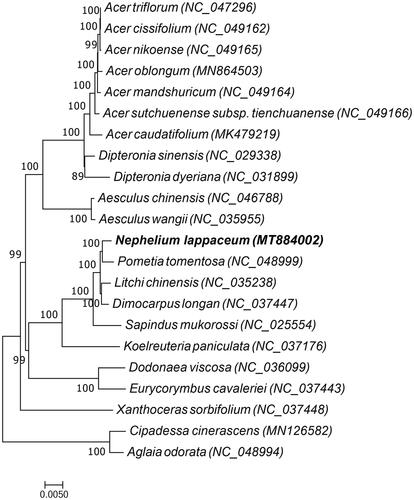
A maximum likelihood phylogenetic tree based on the complete chloroplast genome sequences of 22 plant species was constructed to ascertain the placement of rambutan within the Sapindaceae family. Twenty species belong to the family Sapindaceae, while the two species Aglaia odorata and Cipadessa cinerascens of the Meliaceae family was used as an outgroup. The 22 complete chloroplast genome sequences were aligned by MAFFT (Katoh and Standley Citation2013), and maximum likelihood analysis was performed by RAxMLbased on the GTRGAMMA substitution model (Stamatakis Citation2014) with 1000 bootstrap replicates. The result indicated that compared to other species of the Sapindaceae family, rambutan is more closely related to Pometia tomentosa (). This study can provide a reference for developing markers and further research on the phylogeny and evolution of the Sapindaceae family.
Disclosure statement
The authors confirm that there are no relevant financial or non-financial competing interests to report.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT884002. The associated **BioProject**, **SRA**, and **Bio-Sample** numbers are PRJNA669909, SRR12904123, and SAMN16480967 respectively.
Additional information
Funding
References
- Almeyada N, Mab SE, Martin FW. 1979. The rambutan. Citrus Sub-Trop. Fruit J. 544:10–12.
- Green PS, Popenoe W. 1975. Manual of tropical and subtropical fruits. Kew Bull. 30(4):718.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu J, Niu YF, Ni SB, He XY, Zheng C, Liu ZY, Cai HH, Shi C. 2018. The whole chloroplast genome sequence of Macadamia tetraphylla (Proteaceae). Mitochondrial DNA B Resour. 3(2):1276–1277.
- Mahmood K, Kamilah H, Alias AK, Ariffin F. 2018. Nutritional and therapeutic potentials of rambutan fruit (Nephelium lappaceum L.) and the by-products: a review. J Food Meas Charac 12:1556-1571.
- Nawawi ANN, Hattori M, Kurokawa M, Kurokawa M, Shiraki K. 1999. Inhibitory effects of Indonesian medicinal plants on the infection of herpes simplex virus type 1. Phytother Res. 13(1):37–41.
- Niu YF, Li KX, Liu J. 2020. Complete chloroplast genome sequence and phylogenetic analysis of Annonareticulata. Mitochondrial DNA B Resour. 5(3):3558–3560.
- Nurhuda HH, Maskat MY, Mamot S, Afiq J, Aminah A. 2013. Effect of blanching on enzyme and antioxidant activities of rambutan (nephelium lappaceum) peel. Int Food Res J. 20(4):1725–1730.
- Odey MO. 2012. Comparative anti-nutrients assessment of pulp, seed and rind of rambutan (Nephelium Lappaceum). Ann Bio Res. 3(11):5151–5156.
- Palanisamy U, Cheng HM, Masilamani T, Subramaniam T, Teng LL, Radhakrishnan AK. 2008. Rind of the rambutan, Nephelium lappaceum, a potential source of natural antioxidants. Food Chem. 109(1):54–63.
- Palanisamy UD, Ling LT, Manaharan T, Appleton D. 2011. Rapid isolation of geraniin from Nephelium lappaceum rind waste and its anti-hyperglycemic activity. Food Chem. 127(1):21–27.
- Phang SCW, Palanisamy UD, Kadir KA. 2019. Effects of geraniin (rambutan rind extract) on blood pressure and metabolic parameters in rats fed high-fat diet. J Integr Med. 2:100-106.
- Soeng S, Evacuasiany E, Widowati W, Fauziah N, Manik V, Maesaroh M. 2015. Inhibitory potential of rambutan seeds extract and fractions on adipogenesis in 3T3-L1 cell line. J Exp Integr Med. 5(1):55–60.
- Stamatakis A. 2014. RaxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Thitilertdecha N, Teerawutgulrag A, Kilburn JD, Rakariyatham N. 2010. Identification of major phenolic compounds from Nephelium lappaceum L. and their antioxidant activities. Molecules. 15(3):1453–1465.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wall MM. 2006. Ascorbic acid and mineral composition of longan (Dimocarpus longan), lychee (Litchi chinensis) and rambutan (Nephelium lappaceum) cultivars grown in Hawaii. J. Food Compos Anal. 19(6-7):655–663.
- Yuvakkumar R, Suresh J, Nathanael AJ, Sundrarajan M, Hong SI. 2014. Novel green synthetic strategy to prepare ZnO nanocrystals using rambutan (Nephelium lappaceum L.) peel extract and its antibacterial applications. Mater Sci Eng C Mater Biol Appl. 41:17–27.