Abstract
Lilium rosthornii is the perennial herbaceous bulbous plant belonging to the Lily of the Liliaceae, with high ornamental value and medicinal values. In this present study, we sequenced the complete chloroplast genome of Lilium rosthornii by Illumina Hiseq X Ten and PacBio RS technologies firstly. The genome size of L. rosthornii, was 152,242bp, with typical tetragonal structure: one large single-copy (LSC, 81,875 bp), one small single-copy (SSC, 17,553 bp), and a pair of inverted repeat regions (IRs, 26,407 bp). The overall GC content was 37.02%. The complete genome contained 131 genes, including 85 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Phylogenetic analysis placed L. rosthornii under the family Liliaceae.
Lilium rosthornii belongs to perennial herb of Liliaceae, which is a wild specie and rarely cultivated. It originated from the gullies, streams and forests around Jinfo Mountain, Nanchuan City, Chongqing, with an altitude of 350–1100 m. The flowering period of L. rosthornii is from June to September. Dried fleshy scales are often used as medicinal materials, which has the excellent efficacy in the treatment of lung diseases, health preserving and enriching blood. The L. rosthornii as an perennial herbaceous bulbous plant with high ornamental value (Liu et al Citation2006). It has a long flowering period, the petals are usually reddish yellow or yellow. Besides its flower type feels like Lilium lancifolium, the perianth has purple spots and its texture is smooth.
Vegetative production is an effective method to preserve germplasm and utilize plant traits of L. rosthornii, which is an important resource of Liliaceae biodiversity, at the same time it is a significant breeding material in horticulture (Wang and Sha Citation2008). Due to the large-scale collection, its wild resources have been seriously damaged. The success of tissue culture and rapid propagation system provides technology and way for the protection and sustainable utilization of this wild species. In the present study, we determined the chloroplast genome sequence of Lilium rosthornii, and discussed the genetic relationship among various species in Liliaceae.
The complete genomic DNA was extracted from fresh plant leaves and Lilium rosthornii was collected from the national natural reserve of Hupin mountain in Hunan province. Additional specimens were kept in Herbarium of Yunnan Agricultural University under the collection number 2020WHZ002. Total genomic DNA was isolated from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Valencia, California, USA) according to the manufacturer’s instructions to construction chloroplast DNA libraries. Sequencing was carried out on an Illumina NovaSeq platform. The output was a 5 Gb raw data of 150 bp paired end reads, further trimmed and assembled using SPAdes (Bankevich et al. Citation2012). Resultant clean reads were assembled using GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle). Annotations of chloroplast genome were conducted by the software Geneious (Kearse et al.Citation2012) and checked by comparison against the Lilium taliense complete chloroplast genome (GenBank accession number: KY009938).
The results of whole genome sequencing showed that the size of chloroplast genome of L. rosthornii (Genbank accession number: MW136390) was 152,242 bp with a typical tetragonal structure: one large single-copy (LSC, 81,875 bp), one small single-copy (SSC, 17,553 bp) and two reverse repeats (IRS, 26,407 bp). The total GC content was 37.0%, the GC content of large single copy (LSC) was 34.8%, and the GC content of small single copy (SSC) was 30.6%. A total of 131 genes were detected, including 85 protein-coding genes, 38 tRNA genes and 8 rRNA genes. Nineteen gene are partially or completely duplicated, including seven PCG (rpl2; rpsl23; ycf2; ndhB; rps12; rps7; ycf1), eight tRNA (trnI-GAU, trnA-UGC, trnL-CAA, trnI-CAU, trnR-ACG, trnV-GAC, trnN-GUU, trnH-GUG) and all four rRNA (4.5S, 5S, 16S & 23S rRNA). All the rRNA genes in the genome sequence were located in the repeat region.
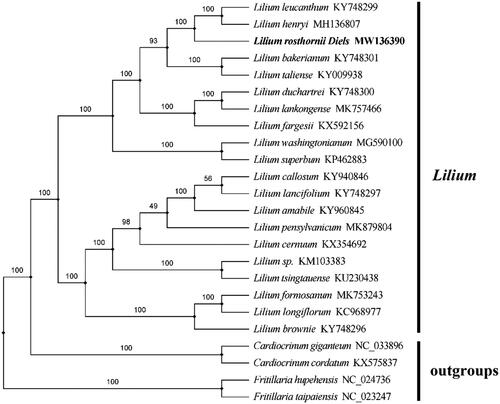
Phylogenetic analysis was constructed based on the complete chloroplast genome sequence obtained from L. rosthornii with those of 25 reported species in the Lilium genus and four out-groups using the maximum likelihood (ML) analysis by RAxML 8.0 software (Stamatakis Citation2014). The maximum likelihood tree was constructed with 1000 bootstrap replicates using FastTree software (Liu et al. Citation2011). According to the result of the analysis, L. rosthornii belongs to family Liliaceae and is associated with other Lilium species. The taxonomic status of L. rosthornii exhibits a closest relationship with L. taliense and L. bakerianum. This finding could serve as valuable genomic resources providing insight into conservation and exploitation efforts for this medicinal species. ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number MW136390.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu YQ, Hu KZ, Xiao B, Xie XM, Liu J. 2006. Studies on tissue culture of Lilium rosthornii. Chin Med Resour. 8(9):32–33.
- Liu K, Randal LC, Tandy W, Rongling W. 2011. RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation. PLOS One. 6(11):e27731.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang YZ, Sha BC. 2008. Tissue culture and rapid propagation of Lilium rosthornii. Subtropical Plant Sci. 37(3):69–70.