Abstract
Spinicaudatan clam shrimp are a widespread and diverse group of branchiopod crustaceans, yet few mitochondrial genomes have been published for this taxonomic group. Here, we present the mitogenome of Leptestheria brevirostris from a rock pool ecosystem in Botswana. Massively parallel sequencing of a single specimen facilitated the reconstruction of the species’ 15,579 bp circularized mitogenome. The reconstructed phylogenetic tree confirms that L. brevirostris forms a monophyletic group with other diplostracan branchiopods, and that these are the sister taxon to Notostraca. The mitogenome reconstructed here is the first to be reported from a leptestherid clam shrimp.
Spinicaudatan clam shrimps are a group of branchiopod crustaceans found in seasonally astatic aquatic habitats on all continents except Antarctica (Brendonck et al. Citation2008; Rogers Citation2009). These shrimps produce resting eggs capable of withstanding prolonged dry conditions between hydroperiods (Brendonck Citation1999). As spinicaudatan clam shrimps tend to occupy and feed in the benthic portions of pools, they are often overlooked (Brendonck et al. Citation2008; Rogers Citation2009). Here, we describe the complete mitogenome of Leptestheria brevirostris Barnard, Citation1924, a leptestherid clam shrimp that was collected from a rock pool temporary wetland in Central District, Botswana. This study forms the basis for more comprehensive phylogenetic studies to better understand the evolution of branchiopod crustaceans and their relatives.
Whole specimens of L. brevirostris were collected from a temporary rock pool (22°35′55.50″ S; 27°7′51.78″ E), preserved in 80% ethanol and identified using the relevant literature (Barnard Citation1924, Citation1929; Brendonck Citation1999). Voucher specimens from the same locality were deposited at the Kansas Biological Survey (DCR-1140). Genomic DNA of high molecular weight was extracted from a single specimen using the CTAB method (Doyle and Doyle Citation1987). A genomic DNA library was constructed from 1 µg of genomic DNA as template using a NEBNext Library Preparation Kit (Ipswich, MA) and sequenced on an Illumina HiSeq 4000 platform using 2 × 150 bp chemistry following the manufacturer’s instructions.
The sequencing run produced 22,933,816 paired-end sequences. The assembly of raw sequences using NovoplastyV4.2 (Dierckxsens et al. Citation2017) resulted in a circular genome with a total length of 15,579 bp. Annotation of the reconstructed mitogenome in MITOS webserver (Bernt et al. Citation2013) identified 13 protein-coding genes, 22 tRNAs, and 2 rRNAs, as is typical of all crustaceans. Instances of non-canonical start codons and truncated stop codons were observed (Jagatap et al. Citation2019; Monsanto et al. Citation2019). The protein-coding DNA sequences of the study species were aligned with those of eight closely related crustaceans in MAFFT v7.429 (Katoh and Standley Citation2013). A consensus Bayesian phylogenetic tree was reconstructed for the study species using BEAST2 v2.6.2 (Bouckaert et al. Citation2014). The program’s default settings were used, except that the substitution model was set to HKY (Hasegawa et al. Citation1985) with four gamma rate categories. Ten independent runs, each comprising, 500 million iterations with 150 million initial burn-in steps were executed in parallel. Final log and tree files were combined in BEAST2 LogCombiner (Rambaut and Drummond Citation2014), and convergence of the independent runs and effective sample size (ESS) were checked in Tracer v1.7 (Rambaut et al. Citation2018). The Bayesian phylogenetic tree was rooted using the anostracans Artemia sinica and Streptocephalus cafer, and visualized in FigTree v1.4 (Rambaut and Drummond Citation2016) ().
Figure 1. A Bayesian phylogenetic tree constructed in BEAST2 using mitogenome sequences of Leptestheria brevirostris (NCBI accession number MN548772) and nine other crustacean species: Limnadia lenticularis (NC_039394.1), Gondwanalimnadia sp. (MN625703.1), Daphnia magna (MK370029.1), Diaphanosoma dubium (NC_037488.1), Lepidurus arcticus (MK579380.1), Triops longicaudatus (KM516710.1), Artemia sinica (MK069595.1), and Streptocephalus cafer (MN720104.1). The numbers on the tree indicate the posterior probability of each node. The scale beneath the tree is expressed in number of substitutions per time unit.
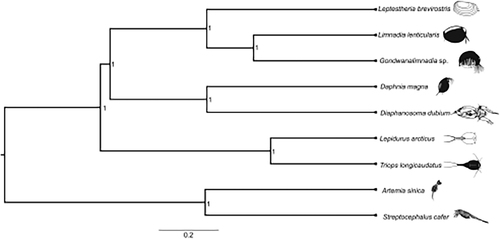
The phylogenetic reconstruction grouped L. brevirostris with other spinicaudatans, and confirmed the monophyly of cladocerans and spinicaudatans and are consistent with previous studies on crustaceans mitogenomics (Luchetti et al. Citation2019) ().
Acknowledgements
The Ministry of Environment, Natural Resources Conservation and Tourism (Botswana) is acknowledged for issuing a research permit (ENT 8/36/4XXXXII(14)). The Centre for High Performance Computing (CHPC) in Cape Town and the University of Johannesburg’s IT services are acknowledged for providing necessary computational platforms and bioinformatics support.
Disclosure statement
The authors declare that there is no existing competition and/or financial interest. Therefore, opinions, findings, conclusions or recommendations expressed in this material are owned by the authors.
Data availability statement
The assembled and annotated mitogenome of the study species is available on the NCBI database (accession number MN548772; https://www.ncbi.nlm.nih.gov/search/all/?term=MN548772).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Barnard KH. 1924. Contributions to a knowledge of the fauna of southwest Africa. II. Crustacea Entomostraca, Phyllopoda. Ann S Afr Mus. 20:213–228.
- Barnard KH. 1929. Contributions to the crustacean fauna of South Africa. 10. Revision of Branchiopoda. Ann S Afr Mus. 29:189–272.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond A. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 10(4):e1003537.
- Brendonck L, Rog Rs DC, Olesen J, Weeks S, Hoeh WR. 2008. Global diversity of large branchiopods (Crustacea: Branchiopoda) in freshwater. Hydrobiologia. 595(1):167–176.
- Brendonck L. 1999. Conchostraca. In: Day JA, Stewart BA, de Moor IJ, Louw AE, editors. Guides to the freshwater invertebrates of Southern Africa. Crustacea I: Notostraca, Anostraca, Conchostraca, and Cladocera. Water Research Commission Report TT121/00; 126 pp.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 22(2):160–174.
- Jagatap H, Monsanto DM, Jansen van Vuuren B, Janion-Scheepers C, Sekar S, Teske PR, Emami-Khoyi A. 2019. The complete mitogenome of the springtail Tullbergia bisetosa: a subterranean springtail from the sub-Antarctic region. Mitochondrial DNA B. 4(1):1594–1596.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Luchetti A, Forni G, Skaist AM, Wheelan BM, Mantovani B. 2019. Mitochondrial genome diversity and evolution in Branchiopoda (Crustacea). Zool Lett. 5:15.
- Monsanto DM, Jansen van Vuuren B, Jagatap H, Jooste CM, Janion-Scheepers C, Teske PR, Emami-Khoyi A. 2019. The complete mitogenome of the springtail Cryptopygus antarcticus travei provides evidence for speciation in the Sub-Antarctic region. Mitochondrial DNA Part B. 4(1):1195–1197.
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 67(5):901–904.
- Rambaut A, Drummond A. 2016. TreeAnnotator v2. 4.5 as part of the BEAST software package. Edinburgh (UK): Institute of Evolutionary Biology, University of Edinburgh.
- Rambaut A, Drummond AJ. 2014. LogCombiner v2. 1.1. Edinburgh (UK): Institute of Evolutionary Biology, University of Edinburgh.
- Rogers DC. 2009. Branchiopoda (Anostraca, Notostraca, Laevicaudata, Spinicaudata, Cyclestherida). In: Likens GE, editor. Encyclopedia of inland waters. Vol. 2. Oxford: Elsevier; p. 242–249.
