Abstract
The complete mitochondrial DNA (mtDNA) for Odorrana exiliversabilisLi, Ye and Fei 2001 (Anura: Ranidae) was determined in this study. The length of complete mtDNA was 17,122 bp, including 13 PCGs (COI-III, ND1-6, ND4L, ATP6, ATP8 and CYTB), 25 tRNA genes, 2 rRNA genes, 2 non-coding regions of a L-strand replication origin and a control region. The overall base composition of the sequence is 28.27% T, 28.27% C, 28.52% A, and 14.94% G, with a total A + T content of 56.79%. The phylogenetic tree showed that O. exiliversabilis was the sister species of O. tormota, and formed a monophyletic clade with other Odorrana species. These data provide a powerful tool for evolutionary biology and population genetics of genus Odorrana.
There are currently fifty-nine species of Odorrana (Anura: Ranidae) distributed in high-gradient streams of Japan, southern China and Indochina west to northeastern India, Myanmar and Thailand and south to through Malaya and Sumatra to Borneo (Frost Citation2020). Thirty-seven species of Odorrana are currently known in China, and twenty-six of which are endemic to China (AmphibiaChina Citation2020). The Fujian Bamboo-leaf Frog (Odorrana exiliversabilis Li, Ye and Fei 2001) is an endemic Chinese lotic frog, which is distributed in Southern Anhui, western Zhejiang, and Fujian Provinces of China (AmphibiaChina Citation2020). In GenBank, seven complete mitochondrial DNA (mtDNA) are available for Odorrana speices, but not including O. exiliversabilis. Here, we present the complete mtDNA sequence of O. exiliversabilis via Illumina sequencing, and found that it had a close relationship with the other species from the genus Odorrana.
We collected the specimen (LSU20200716WY02) of O. exiliversabilis in Mount Wuyi National Park, Nanping, Fujian Province, China (N27.70342°, E117.65675°) in July 2020. Presently, the specimen was stored in the Laboratory of Amphibian Diversity Investigation (ADI) at Lishui University. Total DNA was extracted from muscle tissue of O. exiliversabilis using EasyPure Genomic DNA Kit (TransGen Biotech Co, Beijing, China). The mtDNA was sequenced by Illumina NovaSeq 6000 (Novogene Bioinformatics Technology Co. Ltd., Tianjin, China) for PE 2 × 150 BP sequencing. Raw sequence data (15.57 G) was deposited in NCBI’s Sequence Read Archive (SRA accession: SRR12536539). The NOVO Plasty 3.7 was used to de novo assembled the clean data without sequencing adapters (Dierckxsens et al. Citation2017).
The complete mtDNA sequence (Genbank accession: MT934403) of O. exiliversabilis was 17,122 bp in length, which including 13 PCGs (COX1-3, ND1-6, ND4L, ATP6, ATP8 and CYTB), 25 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 2 non-coding regions of a L-strand replication origin and a control region. The overall base composition of the sequence is 28.27% T, 28.27% C, 28.52% A, and 14.94% G, with a total A + T content of 56.79%. The number of tRNAs in O. exiliversabilis was great than in other Odorrana species (Su et al. Citation2007; Bu et al. Citation2016; Huang et al. Citation2016; Jin et al. Citation2020). There are two duplicate genes of tRNAAla, tRNAAsn, and tRNATyr in O. exiliversabilis mtDNA. All genes were encoded on the H-strand except ND6 and eleven tRNA genes (tRNAPro, tRNAGln, 2 tRNAAla, 2 tRNAAsn, tRNACys, 2 tRNATyr, tRNASer, and tRNAGlu), which were encoded on the L-strand. Among the 13 PCGs, the ND5 was the longest (1,797 bp), while the ATP8 was the shortest (162 bp). Codon usage analysis of O. exiliversabilis showed that four kinds of start codons (ATA, ATG, ATT, and GTG) and six kinds of stop codons (AGA, AGG, TAA, TAG, TA, and T) were used. The 25 tRNA genes varied in size from 63 to 73 bp. The 12S rRNA (938 bp) and 16S rRNA genes (1,585 bp) are located between tRNAPhe and tRNAVal and between tRNAVal and tRNALeu, respectively.
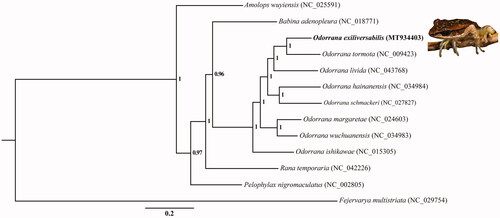
Phylogenetic analysis was inferred from available mtDNA of O. exiliversabilis and other 11 ranid species based on 13 PCGs with Fejervarya multistriata (Anura: Dicroglossidae) as the outgroup using Bayesian inference (BI) methods. The optimal substitution model (GTR + I + G) was implemented by MrModelTest 2.3 (Nylander Citation2004). We analyzed four parallel runs of Markov Chain Monte Carlo (MCMC) for 1,000,000 generations, sampling every 1000 generations and discarded 1000 trees as burn-in. The phylogenetic tree showed that O. exiliversabilis was the sister species of O. tormota, and formed a monophyletic clade with other Odorrana species (). O. ishikawae was a basal clade relative to others within Odorrana, which is similar to the results from Bu et al. (Citation2016) and Jin et al. (Citation2020). These data provide a powerful tool for evolutionary biology, population genetics of genus Odorrana.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Mitogenome data supporting this study is openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MT934403. The SRA accession number is https://www.ncbi.nlm.nih.gov/sra/SRR12536539.
Additional information
Funding
References
- AmphibiaChina. 2020. The database of Chinese amphibians. Kunming, Yunnan, China: Kunming Institute of Zoology (CAS). [accessed 2020 Sep 1]. http://www.amphibiachina.org/.
- Bu XJ, Zhang LJ, He KX, Jiang YM, Nie LW. 2016. The complete mitochondrial genome of the Odorrana schmackeri (Anura, Ranidae). Mitochondrial DNA B. 1(1):162–163.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fei L, Ye CY, Li C. 2001. Taxonomic studies of Odorrana versabilis in China. II. Descriptions of two new species (Amphibia: Ranidae). Acta Zootaxonomica Sinica. 26:601–607.
- Frost DR. 2020. Amphibian species of the world: an online reference, version 6.0. New York (NY): American Museum of Natural History. [accessed 2020 Sep 1]. http://research.amnh.org/herpetology/amphibia/index.php/.
- Huang YJ, Wei Zhao W, Bao XK, Lin YH, Ran JC. 2016. Sequence and analysis of the complete mitochondrial genome of the Wuchuan Odorous Frog Odorrana wuchuanensis (Anura: Ranidae). Mitochondrial DNA B. 1(1):757–758.
- Jin XX, Li WY, Hu SJ, Li WM, Yang JC. 2020. The complete mitochondrial genome of large odorous frog, Odorrana graminea (Amphibia: Ranidae) and phylogenetic analysis. Mitochondrial DNA B. 5:3157–3158.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- Su X, Wu XB, Yan P, Cao SY, Hu YL. 2007. Rearrangement of a mitochondrial tRNA gene of the concave-eared torrent frog, Amolops tormotus. Gene. 394(1–2):25–34.