Abstract
The bovine hookworm Bunostomum phlebotomum (Nematoda: Bunostominae) is a blood-feeding nematode with important socioeconomic impact in the cattle breeding industry. In the present study, the complete mitochondrial genome sequence of a representative individual of B. phlebotomum from beef cattle in Southwest China was determined using the next generation sequencing technology. The genome was 13,799 bp in size and encoded 12 protein-coding genes, 22 tRNA genes and two rRNA genes. The phylogeny revealed that although B. phlebotomum from Chinese beef cattle and yaks were more closely related to each other than to that from Australian cows, these three bovine-originated B. phlebotomum grouped together and formed paraphyletic relationships with Bunostomum trigonocephalum (goat/sheep hookworm) and Necator americanus (human hookworm), supporting their sister-species relationships within Bunostominae. The cumulative mitochondrial DNA data provides a better understanding of phylogenetic relationships of this species in cattle.
The bovine hookworm Bunostomum phlebotomum (Nematoda: Bunostominae) is a blood-feeding nematode that parasitizes the small intestines of the beef cattle, cow, buffalo and yak and cause the socioeconomically important disease ancylostomiasis in the cattle breeding industry (Sprent Citation1946; Huang and Shen Citation2006). Like other hookworms, the infective third-stage larvae (iL3s) of B. phlebotomum can be swallowed or penetrate the skin of hosts and migrate via the blood-circulatory system and lung to finally settle down in the duodenum as dioecious adults. The adults attach to the intestinal mucosa and feed on blood. Consequently, infected animals suffer anemia, weight loss or stunted growth, and even death in cases of heavy infections (Huang and Shen Citation2006; Gao et al. Citation2014). Increased epidemiological surveys suggest that B. phlebotomum has been a worldwide distribution and is emerging as an important pathogen responsible for gastrointestinal infections in cattle (Squire et al. Citation2018; Ola-Fadunsin et al. Citation2020; Charlier et al. Citation2020; Hildreth and McKenzie Citation2020). Unfortunately, until now diagnosis of this hookworm infection has still largely relied on fecal microscopy and often mistaken even by experienced microscopists due to possible environmental cross-contaminating eggs of morphologically similar Bunostomum spp. (Mönnig Citation1950; Wang et al. Citation2012; Gao et al. Citation2014) and possible co-occurring larvae of Haemonchus spp., Strongylus spp. or Chabertia spp (Charlier et al. Citation2020; Hildreth and McKenzie Citation2020). Therefore, obtaining a more efficient and reliable approach to identify and differentiate B. phlebotomum eggs or larvae has become crucial for diagnosis and epidemiological investigation, and achieving this goal is foreseeable only through utilization of molecular methodologies. Mitochondrial DNA (mtDNA)-based PCR is regarded as an efficient molecular tool and has been widely used for species-specific identification and differentiation of many parasitic nematodes of socioeconomic importance (Monis et al. Citation2002; Hu et al. Citation2004; Hu and Gasser Citation2006). To date, there have been two mtDNAs of B. phlebotomum have been characterized with one from cows and another from yaks. However, increased molecular evidences showed that origins (including hosts and geographies) of parasites are capable of shaping cryptic speciation and species diversity, even in one species (Su Citation2014; Korhonen et al. Citation2016), which highlights the significance and necessity to sequence mtDNAs of one species among different hosts and/or geographies. Herein, we sequenced the complete mitochondrial genome sequence of a representative B. phlebotomum from beef cattle in Southwest China and added novel mtDNA data to this hookworm nematode.
The hookworm specimens were obtained from a naturally infected Chinese Simmental beef calf housed in a cattle farm at Yibin (28°09′N, 104°68′E), Sichuan Province of Southwest China, after treatment with pyrantel pamoate. After morphological identification (Mönnig Citation1950) and molecular sequencing the first and second internal transcribed spacers (ITS1 and ITS2) of nuclear ribosomal DNA (Wang et al. Citation2012), all worms (n = 4) were identified as adult females of B. phlebotomum. One worm specimen was used for DNA extraction, and the others were fixed in 5% formalin solution and archived in the Parasitological Museum of Sichuan Agricultural University (Sichuan, China) under collection numbers XY2018_20-22. The genomic DNA was isolated and sequenced using the Illumina HiSeq platform (Novogene, Tianjin, China). The complete mtDNA was assembled using MITObim (Hahn et al. Citation2013) and annotated by MITOS (Bernt et al. Citation2013), as previously described (Xie et al. Citation2019). The complete genome sequence was deposited in GenBank under accession number: MW067147.
The mitochondrial genome of B. phlebotomum was 13,799 bp in size with 77.1% AT and encoded 12 protein-coding genes, 22 tRNA genes and two rRNA genes. All genes were located on the same strand and transcribed in one direction, typical for other hookworms reported so far. Among the 12 protein-coding genes, except cox1, cox3 and nad5 deduced to use an incomplete stop codon ‘T’, the rest were predicted to use the typical TAG (e.g., atp6, nad1, nad4L, nad6 and cytb) or TAA (e.g., cox2 and nad2-4) as the stop codons. Twenty-two tRNA genes ranged from 54 bp (tRNA-Gly, tRNA-His and tRNA(AGN)-Ser) to 62 bp (tRNA-Lys) in size and had nematode-typical stem-loop structures when compared to those of metazoan mtDNAs (Hu and Gasser Citation2006; Jex et al. Citation2009; Gao et al. Citation2014; Xie et al. Citation2019). Both rRNAs, the small rRNA (12S; 694 bp) and large (16S; 961 bp) subunits, were present between tRNA-Glu and tRNA(UCN)-Ser and between tRNA-His and nad3, respectively. Three non-coding regions, namely NC1 (also known as AT-rich region; 235 bp), NC2 (108 bp) and NC3 (24 bp), were located between tRNA-Ala and tRNA-Pro, between nad4 and cox1 and between nad3 and nad5, respectively.
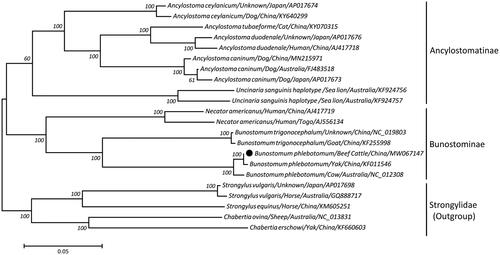
On the basis of a concatenated nucleotide sequence dataset of 12 protein-coding genes from 22 hookworms, a maximum-likelihood (ML)-based phylogeny was reconstructed using species of Strongylidae as outgroup. The phylogenic tree revealed that B. phlebotomum from Chinese beef cattle and yaks were more closely related to each other than to that from Australian cows, nevertheless, these three bovine-originated B. phlebotomum isolates grouped together and formed a branch that was paraphyletic with the congeneric Bunostomum trigonocephalum (goat/sheep hookworm) and Necator americanus (human hookworm) with 100% bootstrap confidence, consistent with previous molecular studies (Jex et al. Citation2009; Gao et al. Citation2014), supporting their sister-species relationships within the family Bunostominae (). Taken together, the sequenced B. phlebotomum mtDNA in this study not only provides novel molecular insights into phylogenetic relationships and taxonomic positions among ruminant hookworms but also becomes useful genetic markers for identification, population genetics and molecular epidemiology of this species.
Figure 1. Maximum likelihood tree inferred from concatenated nucleotide sequences of 12 mt protein-coding genes of B. phlebotomum and other related nematode species, utilizing GTR model and 10,000,000 bootstrap replications (<50% support not shown). The black circle sign represents the species in this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW067147.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Charlier J, Höglund J, Morgan ER, Geldhof P, Vercruysse J, Claerebout E. 2020. Biology and epidemiology of gastrointestinal nematodes in cattle. Vet Clin North Am Food Anim Pract. 36(1):1–15.
- Gao JF, Zhao Q, Liu GH, Zhang Y, Zhang Y, Wang WT, Chang QC, Wang CR, Zhu XQ. 2014. Comparative analyses of the complete mitochondrial genomes of the two ruminant hookworms Bunostomum trigonocephalum and Bunostomum phlebotomum. Gene. 541(2):92–100.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Hildreth MB, McKenzie JB. 2020. Epidemiology and control of gastrointestinal nematodes of cattle in Northern climates. Vet Clin North Am Food Anim Pract. 36(1):59–71.
- Hu M, Chilton NB, Gasser RB. 2004. The mitochondrial genomics of parasitic nematodes of socio-economic importance: recent progress, and implications for population genetics and systematics. Adv Parasitol. 56:134–213.
- Hu M, Gasser RB. 2006. Mitochondrial genomes of parasitic nematodes-progress and perspectives. Trends Parasitol. 22(2):78–84.
- Huang B, Shen J. 2006. Classific atlas of parasites for livestock and poultry in China. Beijing: Chinese Press of Agricultural Science and Technology; p. 323–324.
- Jex AR, Waeschenbach A, Hu M, van Wyk JA, Beveridge I, Littlewood DT, Gasser RB. 2009. The mitochondrial genomes of Ancylostoma caninum and Bunostomum phlebotomum-two hookworms of animal health and zoonotic importance. BMC Genomics. 10:79.
- Korhonen PK, Pozio E, La Rosa G, Chang BC, Koehler AV, Hoberg EP, Boag PR, Tan P, Jex AR, Hofmann A, et al. 2016. Phylogenomic and biogeographic reconstruction of the Trichinella complex. Nat Commun. 7:10513.
- Monis PT, Andrews RH, Saint CP. 2002. Molecular biology techniques in parasite ecology. Int J Parasitol. 32(5):551–562.
- Mönnig HO. 1950. Veterinary helminthology and entomology. 3rd ed. London: Bailliere, Tindall and Cox; p. 196–199.
- Ola-Fadunsin SD, Ganiyu IA, Rabiu M, Hussain K, Sanda IM, Baba AY, Furo NA, Balogun RB. 2020. Helminth infections of great concern among cattle in Nigeria: insight to its prevalence, species diversity, patterns of infections and risk factors. Vet World. 13(2):338–344.
- Sprent JF. 1946. Some observations on the bionomics of Bunostomum phlebotomum, a hookworm of cattle. Parasitology. 37(3–4):202–210.
- Squire SA, Yang R, Robertson I, Ayi I, Squire DS, Ryan U. 2018. Gastrointestinal helminths in farmers and their ruminant livestock from the Coastal Savannah zone of Ghana. Parasitol Res. 117(10):3183–3194.
- Su XZ. 2014. Tracing the geographic origins of Plasmodium falciparum malaria parasites. Pathog Glob Health. 108(6):261–262.
- Wang CR, Gao JF, Zhu XQ, Zhao Q. 2012. Characterization of Bunostomum trigonocephalum and Bunostomum phlebotomum from sheep and cattle by internal transcribed spacers of nuclear ribosomal DNA. Res Vet Sci. 92(1):99–102.
- Xie Y, Xu Z, Zheng Y, Li Y, Liu Y, Wang L, Zhou X, Zuo Z, Gu X, Yang G. 2019. The mitochondrial genome of the dog hookworm Ancylostoma caninum (Nematoda, Ancylostomatidae) from Southwest China. Mitochondrial DNA B Resour. 4(2):3002–3004.
