Abstract
The genus Crematogaster is a diverse group of ants found around the world. We have completed the mitochondrial genome of Crematogaster teranishii, which is the first mitochondrial genome of the genus. The mitochondrial genome is 17,442 bp long and 20.3% in GC ratio, which is similar to those of other ants. It contains 13 protein-coding genes, two ribosomal RNAs, 22 transfer RNAs, and a control region with same gene order to other myrmicine species. The intergenic region between nad3 and trnA was unusually long compared to other ant species. Phylogenetic analysis showed that C. teranishii was closely related to other members of tribe Crematogastrini.
Genus Crematogaster is a hyper diverse group of ants found all over the world, mainly in the warmer tropical regions (Blaimer and Fisher Citation2013). It is the fifth most diverse ant genus covering over 500 species and 270 subspecies (Bolton Citation2012) and is often the dominant members of fauna (Blaimer and Fisher Citation2013). Species of this genus have a flat petiole and its post petiole connected to the dorsal surface of the gaster, which is unique among all other Myrmicinae ants (Blaimer and Fisher Citation2013). Such peculiarities allow the ants to bend their gasters over its bodies, applying venom to apposing creatures, of which its characteristic posture comes the common names of acrobat ants or cocktail ants. Despite these significances, not a single mitochondrial genome (mitogenome) of this genus is available. We completed mitogenome of Crematogaster teranishii, a small arboreal species found in East Asia, as the first mitogenome of genus Crematogaster.
The ants were collected from a colony found in a fallen branch, in the forest of Geoje Island, Republic of Korea (34°48′57.7″N, 128°38′11.3″E). Total DNA was extracted from worker ants using DNeasy Brood & Tissue Kit (QIAGEN, Hilden, Germany). Sequencing library was constructed using Illumina TruSeq Nano DNA Library Preparation Kit (Illumina, San Diego, CA) following manufacturer’s recommendations with around 350-bp DNA fragments. 5.97 Gbp raw sequences obtained from Illumina HiSeqX at Macrogen Inc. (Seoul, Korea), were filtered by Trimmomatic v0.33 (Bolger et al. Citation2014), de novo assembled and confirmed by Velvet v1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17 (Li et al. Citation2009), and SAMtools v1.9 (Song and Liang Citation2013) under the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr; Park et al., in preparation). Geneious R11 v11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotating the assembled mitogenome based on alignments with other ant mitochondrial genomes and MITOS (Bernt et al. Citation2013) was used to double check the annotations. DNA sample and specimen (95% ethanol) were deposited in the InfoBoss Cyber Herbarium (IN; http://herbarium.infoboss.co.kr; J. Park, Voucher number is IB-30011).
The mitochondrial genome of C. teranishii (GenBank accession: MK940828) is 17,442 bp long, and its GC ratio is 20.3%, both well within the range of available myrmicine mitogenomes 15,213 bp in Cardiocondyla obscurior (KX951753; Liu et al. Citation2019) to 19,748 bp in Atta sexdens (MF591717; Barbosa et al. Citation2019) in length and 17.5% in A. texana (MF417380; Barbosa et al. Citation2019) to 23.9% in Wasmannia auropunctata (NC_030541; Duan et al. Citation2016) in GC ratio. It includes 13 protein-coding genes (PCGs), two ribosomal RNAs, 22 transfer RNAs, and an AT-rich non-coding control region and the order of the 37 genes is identical to those of most Myrmicinae species (Babbucci et al. Citation2014; Vieira and Prosdocimi Citation2019). The intergenic region between nad3 and trnA is 1,145 bp long, unusually long compared to other ants: e.g. Pseudomyrmex particeps shows 468 bp as the second longest (BK010384; Vieira and Prosdocimi Citation2019), which is less than half of that of C. teranishii.
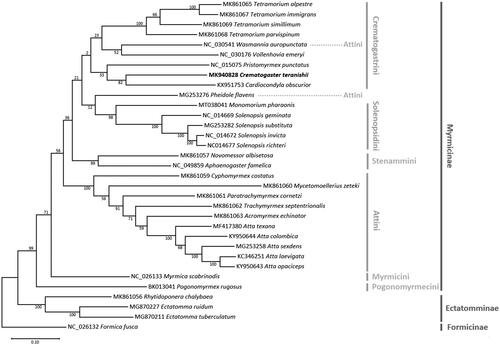
Sequences of 13 PCGs from 29 Myrmicinae and four outgroup mitogenomes were extracted and codon-based alignments were conducted for each PCG using MAFFT provided on the Translator X web server with default settings (Abascal et al. Citation2010). Concatenated alignment was subjected to construct a maximum-likelihood tree using MEGA X (Kumar et al. Citation2018) with default settings. In the phylogenetic tree, tribe Crematogastrini was grouped into one clade with extremely low supportive values (). Relation within the tribe was also chaotic, for instance, C. teranishii directly grouped with Cardiocondyla obscurior (KX951753; ), in contrast to previous studies where Cardiocondyla was a member of the Cataulacus genus group which was suggested to be sister to all other Crematogastrini genera (Ward et al. Citation2015; Blaimer et al. Citation2018). Pristomyrmex punctatus, the species expected to be closest to Crematogaster, instead was sister to the clade of two (). Such weak point could be improved when newly sequenced mitogenomes cover more genera as now only five out of 64 genera have mitogenomes in the tribe. Tribe Attini was also problematic because Wasmannia and Pheidole were closer to tribe Crematogastrini and Solenopsidini, respectively (). The remaining mitogenomes of Attini were from subtribe Attina and occupied the part of the phylogenetic tree displaying congruence to the previous study (Solomon et al. Citation2019). This suggests additional mitogenomes from closely related genera could improve the chaotic status of Crematogastrini clade () since five genera of Crematogastrini are widely dispersed within the tribe, each belonging to separate genus groups (Blaimer et al. Citation2018). Finally, topology among the six tribes was also inconsistent with previous phylogeny (Ward et al. Citation2015). Crematogaster teranishii mitogenome is a valuable addition in our understanding of ant mitochondrial genomes. At the same time, it also presents that more mitogenomes are required in fully understanding inconsistency of phylogenetic relationship of ants.
Figure 1. Maximum-likelihood (bootstrap repeat: 10,000) phylogenetic tree of 29 Myrmicinae ant mitochondrial genomes. The numbers above branches indicate bootstrap support values of the maximum-likelihood tree. Tribe and subfamily names are presented on the right side of the tree with gray and dark gray colors, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MK940828 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA667965, SAMN16392966, and SRR12791370, respectively.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38(Web Server issue):W7–W13.
- Babbucci M, Basso A, Scupola A, Patarnello T, Negrisolo E. 2014. Is it an ant or a butterfly? Convergent evolution in the mitochondrial gene order of Hymenoptera and Lepidoptera. Genome Biol Evol. 6(12):3326–3343.
- Barbosa JT, Barbosa MS, Morais S, Santana AE, Almeida C. 2019. Mitochondrial genomes of genus Atta (Formicidae: Myrmicinae) reveal high gene organization and giant intergenic spacers. Genet Mol Biol. 42(4):e20180055.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Blaimer BB, Fisher BL. 2013. Taxonomy of the Crematogaster degeeri-species-assemblage in the Malagasy region (Hymenoptera: Formicidae). Eur J Taxon. 51:1–64.
- Blaimer BB, Ward PS, Schultz TR, Fisher BL, Brady SG. 2018. Paleotropical diversification dominates the evolution of the hyperdiverse ant tribe Crematogastrini (Hymenoptera: Formicidae). Insect Syst Divers. 2(5):3.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Bolton B. 2012. AntCat. An online catalog of the ants of the world; [accessed 2020 Oct]. http://antcatorg.
- Duan X-Y, Peng X-Y, Qian Z-Q. 2016. The complete mitochondrial genomes of two globally invasive ants, the Argentine ant Linepithema humile and the little fire ant Wasmannia auropunctata. Conserv Genet Resour. 8(3):275–277.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Liu L, Wu Y, Chen F, Wang Q-X, Zhang X-Y, Tang Y, Li F, Qian Z-Q. 2019. Characterization of the complete mitochondrial genome of the invasive tramp ant Cardiocondyla obscurior (Hymenoptera: Formicidae: Myrmicinae). Mitochondrial DNA Part B. 4(1):1496–1498.
- Solomon SE, Rabeling C, Sosa‐Calvo J, Lopes CT, Rodrigues A, Vasconcelos HL, Bacci M Jr, Mueller UG, Schultz TR. 2019. The molecular phylogenetics of Trachymyrmex Forel ants and their fungal cultivars provide insights into the origin and coevolutionary history of ‘higher‐attine’ ant agriculture. Syst Entomol. 44(4):939–956.
- Song N, Liang A-P. 2013. A preliminary molecular phylogeny of planthoppers (Hemiptera: Fulgoroidea) based on nuclear and mitochondrial DNA sequences. PLOS One. 8(3):e58400.
- Vieira GA, Prosdocimi F. 2019. Accessible molecular phylogenomics at no cost: obtaining 14 new mitogenomes for the ant subfamily Pseudomyrmecinae from public data. PeerJ. 7:e6271.
- Ward PS, Brady SG, Fisher BL, Schultz TR. 2015. The evolution of myrmicine ants: phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae). Syst Entomol. 40(1):61–81.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12(Suppl. 14):S2.
