Abstract
The complete mitochondrial genome of Branchinella kugenumaensis from Japan is sequenced, annotated, and compared with that of B. kugenumaensis from China. The mitogenome consists of 14,123 bp with a GC content of 31.83%. Features of both mitogenomes including the presence of regulatory elements in the control region, secondary structure features of tRNAs, and substitution patterns are described and discussed in an evolutionary framework. Comparative studies and genetic analyses indicate high levels of diversity between these two geographically separated populations of B. kugenumaensis, suggesting that they are probably separate species.
Genus Branchinella Sayce 1903 (Anostraca: Thamnocephalidae) is an important member of the macrofaunal community in aquatic environments. These freshwater crustaceans are exceptionally diverse and widespread in Europe, Asia, North and South America, southern Africa, and Australia (Belk and Brtek Citation1995; Brendonck and Belk Citation1997; Remigio et al. Citation2003; Rogers et al. Citation2013). Large scale phylogenies of the Australian Branchinella were reconstructed using a segment of the mitochondrial ribosomal RNA (16S) gene, and the presence of at least three new cryptic species was recovered (Remigio et al. Citation2003; Pinceel et al. Citation2013). Yang and Chen (Citation2020) first published the complete mitogenome of B. kugenumaensis Ishikawa, 1895 from Zhejiang Province, China. Despite the high species diversity, however, mitogenomes of Branchinella remain scarce. In this article, the complete mitogenome of B. kugenumaensis sampled from Japan was determined. A comparative analysis in an evolutionary framework was performed for investigating the evidence of cryptic species at the complete genome level.
Specimens of B. kugenumaensis were collected from a paddy field (134°26′ E, 34°03′ N) near Naruto City, Tokushima Prefecture, Japan. The Voucher specimens (No. BLKNJA01) were deposited in the State Key Laboratory of Palaeobiology and Stratigraphy, Nanjing Institute of Geology and Paleontology, Chinese Academy of Sciences, Nanjing, China. Total DNA was extracted from one adult using the DNeasy tissue kit (Qiagen, Hilden, Germany). The complete mitogenome sequence was determined by long-polymerase chain reaction with conserved primers and sequenced with primer walking using flanking sequences. Overlapped fragments obtained by sequencing were assembled into contigs and aligned using BioEdit 7.0.9.0 (Hall Citation1999). Annotation of mitogenome used MITOS v1 (Bernt et al. Citation2013). Thirteen protein-coding genes were extracted from each mitogenome and aligned using MEGA 7 (Kumar et al. Citation2016). The substitution patterns were obtained using DAMBE 6 version (Xia Citation2017). Bayesian inference (BI) was conducted MrBayes version 3.2 (Ronquist et al. Citation2012). Mean maximum likelihood distances were calculated using MEGA 7 (Kumar et al. Citation2016).
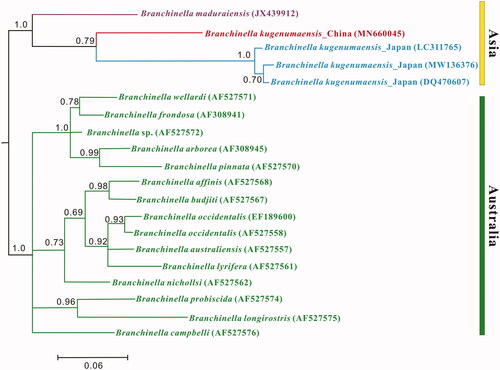
The total length of mitogenome of B. kugenumaensis from Japan (GenBank accession number: MW136376) is 14,123 bp (31.83% GC content), which is 15,127 bp in the Chinese population. This difference is mainly due to the total fraction of noncoding sequences, being 147 and 1182 bp, respectively. Detailed analysis of both genomes shows the presence of peculiar patterns in the control region of B. kugenumaensis from China, e.g. hairpin structures, poly-T tract, AT-rich, and conserved motifs. A total of 94 substitutions occur between the two tRNA sets. The majority of substitutions (48 sites) are observed in the stems. Six PCGs show different lengths between the two genomes: atp8 and nad1 differ by 15 bp, nad5 by 13 bp, atp6 by 11 bp, nad2 by 8 bp, and nad4L by 6 bp. Percent sequence divergence between the two genomes calculated for each aligned PCG ranges from 16.36% (cox1) to 32.05% (atp8) and total number of nonsynonymous codon substitutions ranges from 5 (cox2) to 87(nad5), which constitutes a high level of diversity between the two B. kugenumaensis populations. The unusually high variation observed in the mitogenomes of two geographically separated populations prompted the question whether or not Asian B. kugenumaensis were separate species. Considering the scarce number of mitogenomes, the 16S rRNA gene was utilized to construct the phylogeny of genus Branchinella, and to calculate the genetic distances. The inferred phylogenetic tree based on the 16S rRNA sequences places B. kugenumaensis of Japan within the Asian Branchinella clade, as the sister to B. kugenumaensis of China. The phylogenetic relationships within Australian species are consistent with the topologies of Remigio et al. (Citation2003) and Pinceel et al. (Citation2013). ML-corrected genetic distances within 16 16S rRNA sequences of Branchinella range from 2.9 to 22.8%. The overall average divergence among the 16S rRNA of the 16 Branchinella species in this work is 11.9 ± 2.1%, while the two populations of B. kugenumaensis exhibit high genetic distance: 12.8 ± 2.3%. Similar levels of divergence are also found in comparisons performed on the cox1 bar-coding fragment. To sum up, comparative analyses recover high levels of diversity between two geographically separated populations of B. kugenumaensis. Therefore, it is reasonable to suggest that these two populations of B. kugenumaensis probably belong to two different cryptic species, but this hypothesis needs to be thoroughly tested by more sampling from Asia coupled with more molecular data assessment and morphological characteristics to define appropriate conservation units.
Disclosure statement
No potential conflict of interest was reported by the author.
Data availability statement
All relevant data can be obtained free of charge on NCBI through GeneBank No. MW136376 (NCBI https://www.ncbi.nlm.nih.gov/nuccore/MW136376).
Additional information
Funding
References
- Belk D, Brtek J. 1995. Checklist of the Anostraca. Hydrobiologia. 298(1-3):315–353.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Brendonck L, Belk D. 1997. Branchinella maduraiensis Raj (Crustacean, Branchiopoda, Anostraca) shown by new evidence to be a valid species. Hydrobiologia. 359(1/3):93–99.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Sympos Series. 41(41):95–98.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Pinceel T, Vanschoenwinkel B, Waterkeyn A, Vanhove M, Pinder A, Timms B, Brendonck L. 2013. Fairy shrimps in distress: a molecular taxonomic review of the diverse fairy shrimp genus Branchinella (Anostraca: Thamnocephalidae) in Australia in the light of ongoing environmental change. Hydrobiologia. 700(1):313–327.
- Remigio EA, Timms BV, Hebert PDN. 2003. Phylogenetic systematics of the Australian fairy shrimp genus Branchinella based on mitochondrial DNA sequences. J Crust Biol. 23(2):436–442.
- Rogers DC, Shu S, Yang J. 2013. The identity of Branchinella yunnanensis Shen, 1949, with a brief review of the subgenus Branchinellites (Branchiopoda: Anostraca: Thamnocephalidae). J Crust Biol. 33(4):576–581.
- Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Xia X. 2017. DAMBE6: new tools for microbial genomics, phylogenetics, and molecular evolution. J Hered. 108(4):431–437.
- Yang RS, Chen YT. 2020. The complete mitochondrial genome of the freshwater fairy shrimp Branchinella kugenumaensis Ishikawa 1894 (Crustacea: Anostraca: Thamnocephalidae). Mitochondrial DNA Part B. 5(1):1048–1049.