Abstract
Galega officinalis L. is a perennial herb of the Fabaceae family. The flowers of G. officinalis L. are colorful and suitable for ornamental purposes. It can be used as a food complement for animals and humans, and it could promote lactation in animals and humans. In this study, we obtained the complete chloroplast genome of G. officinalis L. and found it is 125,086 bp in length. The GC content of this genome is 34.18%. Among the 112 unique genes in the chloroplast genome of G. officinalis L., 30 tRNA, 4 rRNA and 78 protein-coding genes were successfully annotated. We constructed the maximum likelihood (ML) tree with 26 species, and concluded that G. officinalis is phylogenetically closely related to the genus of Cicer, Glycine and Desmodium.
Galega officinalis L. is a leguminous perennial herb, which is native to southern Europe and southwest Asia (Oubre et al. Citation1997). It can be used as animal feed, herbal medicine and ornamental plant, and also used to treat diabetes during medieval times (Atanasov and Tchorbanov Citation2002). In particular, G. officinalis L. can increase lactation in humans and animals (Leporatti and Ivancheva Citation2003). However, few studies have focused on the molecular aspects of G. officinalis L.
Chloroplast is an important organelle with its own genome. Studies on chloroplast genome has been a hot topic in plant molecular evolution and systematics (Clegg et al. Citation1994). Several features of chloroplast genome have facilitated molecular evolutionary analyses. However, the chloroplast genome sequences of G. officinalis L. have not been reported so far. In the present study, the chloroplast genome of G. officinalis L. was sequenced and its structural features were characterized, which will be a valuable resource for in-depth studies of the genetic evolution within the Fabaceae family.
Seeds of G. officinalis L. were kept at the Forage Germplasm Bank at Institute of Animal Sciences of the Chinese Academy of Agricultural Sciences (IAS-CAAS) (Beijing, E116°29′, N40°03′). The voucher specimen (FR004) was deposited at the Herbarium of the Department of Grassland, IAS-CAAS, Beijing, China. After germinating in the laboratory, genomic DNA from young leaves was extracted using a DNA Extraction Kit from Tiangen Bio Tech Co., Ltd (Beijing, China). The sequencing was carried out on the Illumina Novaseq PE150 platform (Illumina Inc, San Diego), and 150 bp paired-end reads were generated. The software GetOrganelle v1.5 (Jin et al. Citation2019) was used to assemble the cleaned reads into a complete chloroplast genome, with the chloroplast genome of Medicago truncatula (GenBank accession number: NC_003119) used as a reference. The chloroplast genome annotation was performed through the online program CPGAVAS2 (Shi et al. Citation2019) and GeSeq (Tillich et al. Citation2017), followed by manual correction. The assembled chloroplast genome sequence with annotation was submitted to GenBank database under the accession number MT506239.
In the present study, we found the complete chloroplast genome of G. officinalis L. is 125,086 bp in length, which is a typical circular structure. The GC content of its genome is 34.18%. The chloroplast genome has in total 112 genes, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Among them, 30 genes encoding amino acid transfer protein, 21 genes encoding ribosomal structural proteins protein, 17 genes encoding electron transport protein, 14 genes encoding light collection structural protein (PSII) are found in the chloroplast genome of G. officinalis L. ().
Table 1. Gene predicted of the chloroplast genome of Galega officinalis L.
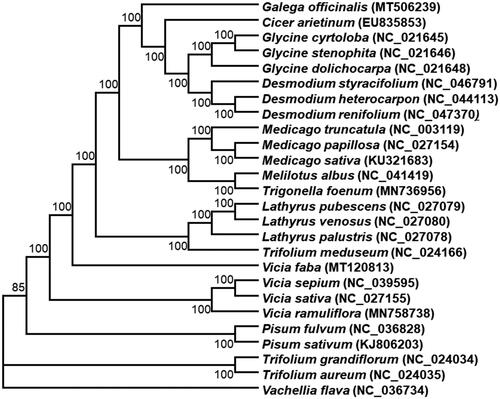
The chloroplast genomes of 26 species from Fabaceae as well as Vachellia flava as outgroup species was downloaded from the NCBI GenBank database to inference the phylogenetic relationship of G. officinalis L. The sequences were aligned using MAFFT v7 (Katoh et al. Citation2019). In addition, a maximum likelihood (ML) tree based on the common protein-coding genes of 26 species was constructed by using IQ-TREE under the model automatically selected by IQ-TREE (Nguyen et al. Citation2015). Phylogenetic analysis shows that G. officinalis L is the sister clade of the clade containing species of Cicer, Glycine and Desmodium (). It was noticed that the Galega chloroplast genome lacks the inverted repeats (IR) as Cicer arietinum, but this atypical plastome architecture can be found in some other clades of the Fabaceae family, including Glycine max and Lotus japonicus (Wang et al. Citation2017). Finally, this study will provide important information for species identification, molecular evolutionary and phylogenetic relationship at different taxonomic levels within the Fabaceae family.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in NCBI at Genbank with accession number MT506239 (https://www.ncbi.nlm.nih.gov/nuccore/MT506239) Raw sequencing reads used in this study was deposited in the public repository SRA with accession number SRR12678613 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR12678613).
Additional information
Funding
References
- Atanasov AT, Tchorbanov B. 2002. Anti-platelet fraction from Galega officinalis L. inhibits platelet aggregation. J Med Food. 5(4):229–234.
- Clegg MT, Gaut BS, Learn GH, Morton BR. 1994. Rates and patterns of chloroplast DNA evolution. Proc Natl Acad Sci USA. 91(15):6795–6801.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. :256479.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Leporatti ML, Ivancheva S. 2003. Preliminary comparative analysis of medicinal plants used in the traditional medicine of Bulgaria and Italy. J Ethnopharmacol. 87(2–3):123–142.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Oubre AY, Carlson TJ, King SR, Reaven GM. 1997. From plant to patient: an ethnomedical approach to the identification of new drugs for the treatment of NIDDM. Diabetologia. 40(5):614–617.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–73.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang YH, Jian QX, Chen SY, Li DZ, Yi TS. 2017. Plastomes of Mimosoideae: structural and size variation, sequence divergence, and phylogenetic implication. Tree Genet Genom. 13:41.