Abstract
The family Trichoceridae is a small group in Nematocera (Diptera) with 157 known species in the world. In this study, we report a complete mitochondrial genome sequence of a Trichoceridae species, which is a circular molecule of 16,094 bp with 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and a 1063 bp long non-coding region (control region). Eight gene overlaps ranging from 1 to 8 bp in length and 16 small non-coding intergenic spacers ranging from 1 to 77 bp in length are found in the mitochondrial genome. The canonical start codons (ATN) for invertebrate mitochondrial genomes are found in 11 PCGs, except for COI which uses TTG and ND5 uses GTG. Stop codons of 12 PCGs are invariably complete TAA and TAG, while COII ends with a single thymine stop codon. Twenty-two tRNAs ranges from 64 bp to 72 bp, and the rRNAs are determined to be 1326 bp in length for lrRNA and 788 bp in length for srRNA. Phylogenetic analysis reveals that the family Trichoceridae is a sister-group to the remaining Tipulomorpha, and the relationship between the other families is Pediciidae + (Limoniidae + (Cylindrotomidae + Tipulidae)).
Introduction
The family Trichoceridae, commonly called the winter crane fly, is a small group in Nematocera (Diptera) with 157 known species (Krzemińska et al. Citation2009). The family is divided into six recent genera within two subfamilies: Diazosma Bergroth, Citation1913, Nothotrichocera Alexander, Citation1926 and Trichocera Meigen, Citation1803 referable to the subfamily Trichocerinae; Adura Krzemińska, Citation2006, Paracladura Brunetti, Citation1911 and Zedura Krzemińska, Citation2005 referable to the subfamily Paracladurinae (Krzemińska Citation1992, Citation2005, Citation2006).
Trichocera Meigen is the largest genus in the winter crane fly with more than 110 known species in the world, of which six species are known from China (Alexander Citation1930, Citation1933, Citation1935a, Citation1935b, Citation1938; Yang and Yang Citation1995). It differs from the other genera by the eye having hairs between the ommatidia, the tibial spurs being present, the first tarsi being longer than the second, the vein A2 being short and curved evenly to the margin of the wing, the crossvein m-cu being absent, and the cerci being downward-curved, elongate and sclerotized (Alexander Citation1981). Two mitochondrial genomes of Trichoceridae have been published (Beckenbach Citation2012), one of which belongs to the genus Trichocera. In this study, another Trichocera mitochondrial genome is sequenced and analyzed, providing insights into the phylogeny of the infraorder Tipulomorpha.
Materials and methods
The specimen of Trichocera sp. used in this study was collected from Linggongli, Mount Emei, Sichuan, China (29°34'43''N, 103°17'29''E; 1300 m) and stored in the Entomological Museum of Qingdao Agricultural University, China (No. TRI0001). The total genomic DNA was extracted from thoracic muscle of the specimen using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China). The fragments of mitochondrial genome were amplified using standard primers for insects (Simon et al. Citation1994) and were assembled using MEGA7 (Kumar et al. Citation2016). The complete sequence was annotated by MITOS WebServer (Bernt et al. Citation2013) with checking for predicted tRNA and protein-coding genes. Phylogenetic tree was reconstructed using the Maximum Likelihood algorithm in MEGA7 software (Kumar et al. Citation2016).
Results and discussion
The complete mitochondrial genome of Trichocera sp. (GenBank accession no. MW263048) is 16,094 bp in length. It contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs) and a 1063 bp long non-coding region (control region) between the srRNA and tRNAlle-tRNAGln-tRNAMet (IQM). The gene order retains the ancestral arrangement for the order Diptera (Clary and Wolstenholme Citation1985), encoding 23 genes on the majority strand and 14 genes on the minority strand. Eight gene overlaps are found in the mitochondrial genome, ranging from 1 to 8 bp in length. The longest overlaps exist between tRNATrp and tRNACys. Sixteen small non-coding intergenic spacers are found in the mitochondrial genome, ranging from 1 to 77 bp in length. The largest non-coding intergenic spacer is between tRNAArg and tRNAAsn. The canonical start codons (ATN) for invertebrate mitochondrial genomes (Wolstenholme Citation1992) are found in 11 PCGs of the mitochondrial genome, while COI and ND5 use uncanonical start codons TTG and GTG respectively. The complete stop codons, TAA and TAG, are found in 12 PCGs of the mitochondrial genome, except for COII using a single thymine stop codon. The entire 22 typical cloverleaf tRNAs in the arthropod mitochondrial genomes are found in the mitochondrial genome, ranging from 64 bp to 72 bp. Most of the tRNAs could be folded into the classic clover-leaf structures, while the DHU arm of the tRNASer(AGN) gene did not form a stable stem-loop structure. The rRNAs in mitochondrial genome are determined to be 1326 bp in length for lrRNA and 788 bp in length for srRNA.
The nucleotide composition of Trichocera sp. mitochondrial genome is biased toward A and T. The overall AT content of the mitochondrial genome is 75.4% (A: 37.8%; T: 37.6%; C: 14.2%; G: 10.4%). For PCGs, the AT content of the N strand genes (75.8%) is higher than that of the J strand genes (71.5%). For rRNAs, the AT content of the lrRNA (80.9%) is slightly higher than that of the srRNA (77.9%). The AT content of the tRNAs is 75.0%. The control region has the highest AT content (88.2%), which is typical of animal mitochondrial genome. The mitochondrial genome has a weakly positive AT-skew and a negative GC-skew on the J-strand. Comparisons of mitochondrial genomes between Trichocera sp. and T. bimacula are listed in . The proportionate number of nonsynonymous differences (dN) is usually divided by the proportionate number of synonymous differences (dS) to quantify the gene evolutionary rates. The resulting ratio (dN/dS) among Trichocera flies () shows that ATP8 has the fastest evolutionary rate, while COI has the slowest evolutionary rate.
Figure 1. The resulting ratio (dN/dS) of 13 protein-coding genes (PCGs) of mitochondrial genomes among Trichocera spp.
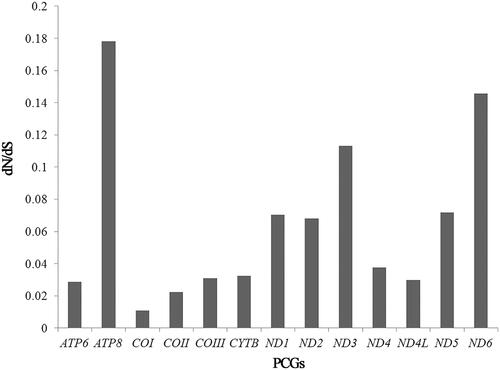
Table 1. Comparisons of mitochondrial genomes between two Trichocera flies.
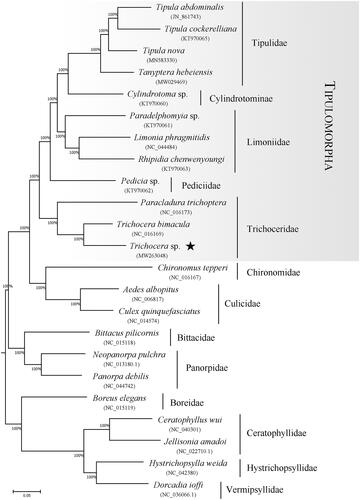
The arrangement of the infraorder Tipulomorpha, containing the families Cylindrotomidae, Limoniidae, Pediciidae, Tipulidae and Trichoceridae, was accepted by Bertone et al. (Citation2008), Dahl (Citation1980), Griffiths (Citation1990), Oosterbroek and Courtney (Citation1995) and Starý (Citation1992). However, the interfamilial relationships in Tipulomorpha are unresolved for a long time. The phylogenetic tree in our study () indicates that the family Trichoceridae is sister-group to the remaining Tipulomorpha, which was accepted by Bertone et al. (Citation2008), Hennig (Citation1973), Kang et al. (Citation2017), Oosterbroek and Courtney (Citation1995) and Zhang et al. (Citation2016). The relationship between the four families of the superfamily Tipuloidea is as follows: Pediciidae + (Limoniidae + (Cylindrotomidae + Tipulidae)).
Acknowledgments
The authors express sincere thanks to Ding Yang (Beijing) and Fan Song (Beijing) for their great help during the study. We are also very grateful to Junchao Wang (Zhengzhou) for collecting the specimens.
Disclosure statement
No potential conflict of interest was report by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW263048. The associated BioProject and Bio-Sample numbers are PRJNA681477 and SAMN16954468, respectively.
Additional information
Funding
References
- Alexander CP. 1926. The Trichoceridae of Australia (Diptera). Proc Linn Soc New South Wales. 60:298–304.
- Alexander CP. 1930. Records and descriptions of Trichoceridae from the Japanese Empire (Ord. Diptera). Konowia. Zeitschrift Fur Systematische Insektenkunde. 9:103–108.
- Alexander CP. 1933. New or little-known Tipulidae from eastern Asia (Diptera). Philipp J Sci. 50:129–162.
- Alexander CP. 1935a. New or little-known Tipulidae from eastern Asia (Diptera). Philipp J Sci. 55:133–164.
- Alexander CP. 1935b. New or little-known Tipulidae from eastern Asia (Diptera). Philipp J Sci. 56:339–372.
- Alexander CP. 1938. New or little-known Tipulidae from eastern Asia (Diptera). Philipp J Sci. 66:309–342.
- Alexander CP. 1981. Trichoceridae. In: McAlpine JF, Peterson BV, Shewell GE, Teskey HJ, Vockeroth JR, Wood DM, editors. Manual of Nearctic Diptera. Vol. I. Ottawa: Research Branch, Agriculture Canada; p. 301–304.
- Beckenbach AT. 2012. Mitochondrial genome sequences of Nematocera (Lower Diptera): evidence of rearrangement following a complete genome duplication in a winter crane fly. Genome Biol Evol. 4(2):89–101.
- Bergroth E. 1913. A new genus of Tipulidae from Turkestan, with notes on other forms. Ann Mag Nat Hist. 11(66):575–584.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bertone MA, Courtney GW, Wiegmann BM. 2008. Phylogenetics and temporal diversification of the earliest true flies (Insecta: Diptera) based on multiple nuclear genes. Syst Entomol. 33(4):668–687.
- Brunetti E. 1911. Revision of the Oriental Tipulidae with descriptions of new species. Record India Museum. 6:231–314.
- Clary DO, Wolstenholme DR. 1985. The ribosomal RNA genes of Drosophila mitochondrial DNA. Nucleic Acids Res. 13(11):4029–4045.
- Dahl C. 1980. Comparison of postembryonic organization of the genital segments Trichoceridae, Tipulidae, and Anisopodidae (Diptera, Nematocera). Zool Scripta. 9(1-4):165–185.
- Griffiths GCD. 1990. Book review: manual of Nearctic Diptera Volume 3. Quaestiones Entomologicae. 26:117–130.
- Hennig W. 1973. Diptera (Zweiflügler). In: Helmcke JG, Starck D, Wermuth H, editors. Handbuch der Zoologie, Bd. 4: Arthropoda, 2. Hälfter: Insecta, 2. Aufl., 2 Teil Spezielles. Berlin: Walter de Gruyter.
- Kang Z, Zhang X, Ding S, Tang C, Wang Y, de Jong H, Cameron SL, Wang M, Yang D. 2017. Transcriptomes of three species of Tipuloidea (Diptera, Tipulomorpha) and implications for phylogeny of Tipulomorpha. PLoS One. 12(3):e0173207
- Krzemińska E. 1992. Paracladurinae-new subfamily (Diptera: Trichoceridae). Acta Zoologica Cracoviensia. 35(1):73–78.
- Krzemińska E. 2005. Subfamily Paracladurinae. III. Phylogenetic biogeography: two new genera and three species described (Diptera, Trichoceridae). N Z J Zool. 32(4):317–352.
- Krzemińska E. 2006. A new species of Nothotrichocera from New Zealand, and a replacement name for the genus Adura (Diptera: Trichoceridae). N Z J Zool. 33(3):229–231.
- Krzemińska E, Krzemiński W, Dahl C. 2009. Monograph of fossil Trichoceridae (Diptera). Over 180 million years of evolution. Kraków: Institute of Systematics and Evolution of Animals, Polish Academy of Sciences.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Meigen JW. 1803. Versuch einer neuen Gattungseinteilung der europäischen zweiflügeligen. Insekten. Magazin Für Insektenkunde. 2:259–281.
- Oosterbroek P, Courtney G. 1995. Phylogeny of the nematocerous families of Diptera (Insecta). Zool J Linn Soc. 115(3):267–311.
- Simon C, Frati F, Beckenbach AT, Crespi B, Liu H, Flook P. 1994. Evolution, weighting and Phylogenetics utility of mitochondrial gene sequences and compilation of conserved polymerase chain reaction Primers. Ann Entomol Soc Am. 87(6):651–701.
- Starý J. 1992. Phylogeny and classification of Tipulomorpha, with special emphasis on the family Limoniidae. Acta Zoologica Cracoviensia. 35:11–36.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Yang J, Yang D. 1995. Diptera: Trichoceridae. In: Zhu Y, editor. Insects and macrofungi of Gutianshan, Zhejiang. Hangzhou: Zhejiang Science and Technology Press; p. 175–179.
- Zhang X, Kang Z, Mao M, Li X, Cameron SL, de Jong H, Wang M, Yang D. 2016. Comparative Mt Genomics of the Tipuloidea (Diptera: Nematocera: Tipulomorpha) and its implications for the phylogeny of the Tipulomorpha. PLoS One. 11(6):e0158167.