Abstract
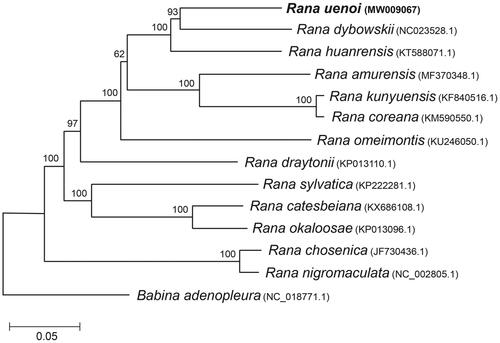
We determined the complete mitochondrial genome of Rana uenoi (Anura: Ranidae) for the first time. The whole sequences were 17,370 bp and included 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. The gene arrangement was completely identical to those observed from other Ranidae species. We used 11 protein-coding genes to examine the phylogenetic placement of this species in the genus Rana. Rana dybowskii was the closest sister species to R. uenoi. The clade of R. uenoi and R. dybowskii formed a cluster with Rana huarensis, which had a sister relationship with the group of Rana amurensis, Rana coreana, and Rana kunyuensis.
Rana uenoi is a species of brown frogs first reported in 2014. This species has previously been assigned to Rana dybowskii (the East Asian brown frog) that was widely distributed across Northeast China and the Far East Russia, Korean Peninsula, and Japan (Yang et al. Citation2017). The taxonomic controversy has long persisted regarding the complex morphological diversity of R. dybowskii, and a series of studies have suggested that R. dybowskii may be a species complex representing multiple species candidates (Matsui et al. Citation1998; Sumida et al. Citation2003; Yang et al. Citation2017). Rana uenoi was officially recorded based on evidence that populations on the Korean Peninsula and Tsushima Island showed a significant differentiation in mtDNA sequence and allozyme characteristics from individuals collected at the type locality (Vladivostok, Russia; Günther Citation1876) of R. dybowskii (Matsui Citation2014). This species is also clearly distinguished from the other two morphologically similar species living on the Korean Peninsula. For example, unlike R. huanrenensis, R. uenoi has paired internal vocal sacs (Matsui Citation2014), and the supralabial lines that R. coreana has distinctly are not observed in this species (Lee and Park Citation2016). The mitochondrial genome sequence of R. uenoi has not been reported to date. However, this data is truly necessary, in that questions still remain regarding the diversity and phylogenetic structure of R. dybowskii species complex (Yang et al. Citation2017).
Here, we analyzed the complete mitochondrial genome sequence of R. uenoi. The DNA sample was isolated from an ethanol-immersed specimen stored in the Conservation Genome Resource Bank for Korean Wildlife (CGRB: http://www.cgrb.org/) that was collected (N36°32′24.1′′/E127°50′50.01′′) in 2016. The specimen is accessible as mms7611 in CGRB. Mitogenomic sequences were extracted using the CLC genomics workbench 6.5 (http://www.qiagenbioinformatics.com/) from the reads generated from the MiSeq platform. Each mitochondrial region was annotated using the MITOS web server (Bernt et al. Citation2013) and manually checked using the mitochondrial information of R. dybowskii (NC023528.1; Li et al. Citation2016). The sequence information was deposited at NCBI GenBank under the accession number of MW009067. The whole sequences were 17,370 bp and included 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. L-strand was observed in eight tRNA genes and ND6. Every protein-coding gene contained ATG start codon with exceptions in ND1 (GTG), COX1 (ATA), and ND4L (GTG). ND2 (TAG), COX1 (AGG), APT8 (TAA), ND4L (TAA), ND6 (AGA), and Cyt b (TAA) were terminated with the stop codons of vertebrate mitochondrial DNA. An incomplete stop codon was detected at the remaining six coding genes. The gene arrangement was completely identical to those observed from R. dybowskii (Li et al. Citation2016). We used 11 protein-coding genes (8964 bp; excluding ND5 and ND6 to avoid incomplete alignment) to examine the phylogenetic placement of this species in the genus Rana (). Rana dybowskii was the phylogenetically closest sister species to R. uenoi. The clade of R. uenoi and R. dybowskii formed a cluster with Rana huarensis, which had a sister relationship with the group of Rana amurensis, Rana coreana, and Rana kunyuensis (). Our study can provide useful information to reconstruct the consensus phylogenetic tree that is essential for the future study of brown frog diversity.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW009067. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA689962, SRR13364873, and SAMN17222395, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Günther ACLG. 1876. Description of a new frog from north-eastern Asia. Ann Mag Nat Hist Ser. 17(101):387.
- Lee J, Park D. 2016. The encyclopedia of Korean amphibians. Seoul (South Korea): Econature.
- Li J, Lei G, Fu C. 2016. Complete mitochondrial genomes of two brown frogs, Rana dybowskii and Rana cf. chensinensis (Anura: Ranidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):155–156.
- Matsui M. 2014. Description of a new brown frog from Tsushima Island, Japan (Anura: Ranidae: Rana). Zool Sci. 31(9):613–620.
- Matsui M, Tanaka-Ueno T, Paik NK, Yang SY, Takenaka O. 1998. Phylogenetic relationships among local populations of Rana dybowskii assessed by mitochondrial cytochrome b gene sequences. Jap J Herpetol. 17(4):145–151.
- Sumida M, Ueda H, Nishioka M. 2003. Reproductive isolating mechanisms and molecular phylogenetic relationships among Palearctic and Oriental brown frogs. Zoolog Sci. 20(5):567–580.
- Yang BT, Zhou Y, Min MS, Matsui M, Dong BJ, Li PP, Fong JJ. 2017. Diversity and phylogeography of Northeast Asian brown frogs allied to Rana dybowskii (Anura, Ranidae). Mol Phylogenet Evol. 112:148–157.