Abstract
Chimonobambusa hejiangensis is a kind of bamboo that has excellent edible and economic value, which is endemic to southwest China. The study used next-generation sequencing to obtain the complete chloroplast (cp) genome sequence of C. hejiangensis. The cp genome of C. hejiangensis has a total length of 138,908 bp, and consisted of an 82,495-bp large single-copy region, an 12,743-bp small single-copy region, and two 21,835-bp IR regions. In total, 112 unique genes were found in the cp genome, including 77 protein coding, 31 tRNA, and 4 rRNA genes. Phylogenetic analysis indicated that C.hejiangensis and C. tumidissinoda are sister species within the Arundinarieae genus, where Chimonocalamus and Ampelocalamus are more closely related to them.
Chimonobambusa hejiangensis C. D. Chu & C. S. Chao is mainly distributed in southern China at the junction of Guizhou and Sichuan provinces. It is suitable for growing in acidic and neutral soils with an optimal altitude of 180–1300 m (Liu et al. Citation2012). Previous research has shown that bamboo shoots from C. hejiangensis excel in autumn, when they are not only delicious but also contain various trace elements such as iron, sun, and zinc, as well as being rich in other nutrients (Gou et al. Citation2010).
Chloroplasts are organelles that take part in photosynthesis and known as the ‘nutrient manufacturing workshop’ and ‘energy conversion station’ in plants. From a structural point of view, the two inverted repeats (IRs) of the chloroplast genome are separated by a large single-copy region (LSC) and a small single copy region (SSC), thus forming a typical quadripartite structure. Compared with two other genetic materials, namely, nuclear genome and linear chloroplast genome, the chloroplast genome has characteristics of strong conservatism, easy access to sequences, and moderate rate of evolution, providing sufficient resources for plant systematics (Wu et al. Citation2009). Related research from morphological identification to the molecular perspective is of great significance in understanding the classification and evolutionary relationships among bamboo species (Zhang et al. Citation2011; Liu et al. Citation2018). In recent years, with the emergence of next-generation sequencing (NGS) technology, chloroplast genome sequences have been progressively analyzed, contributing insight into the phylogenetic traits of plants, transgenes, and plant identification (Ceccherini et al. Citation2003; Wambugu et al. Citation2015; Xue et al. Citation2019).
The research materials were collected in Bao yuan Township, Guizhou Province (28°20′14″N and 105°38′32″E). The voucher specimens of C. hejiangensis were deposited at the Natural Museum of Guizhou University (accession number: GACP). The chloroplast DNA was extracted using a TIANGEN DNA extraction kit (TIANGEN BIOTECH CO., Beijing China) and sequenced based on the Illumina Hiseq 2500 platform (San Diego, CA). Approximately 3GB of paired-end (150 bp) raw short sequence data were generated. The GetOrganelle (Jin et al. Citation2020) was used to de novo assemble the complete cp genome. PGA (https://github.com/quxiaojian/PGA) annotated the chloroplast genes of C. hejiangensis with C. tumidissinoda as the reference followed by manual correction using the software Generous 10.0.5 (Kearse et al. Citation2012), with uploading to NCBI after the sequence was confirmed to be correct (GenBank MT884004).
The chloroplast genome of C. hejiangensis with a length of 138,908 bp, and consisted of an 82,495-bp LSC region, a 12,743-bp SSC region, and two 21,835-bp IR regions, respectively, comprising the typical quadripartite structure of terrestrial plants. The total GC content found for the C. hejiangensis cp genome was 38.93%, and the LSC region (37%) and SSC region (33.2%) have much lower values than that in the IR region (44.22%). The cp genome of C. hejiangensis was annotated with 112 genes, including 77 protein coding, 31 tRNA, and four rRNA genes. Thirteen genes contain one intron (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rps16, trnA-UGC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) and only gene cyf3 includes two introns, while the intron of the gene clpP was found to be deleted.
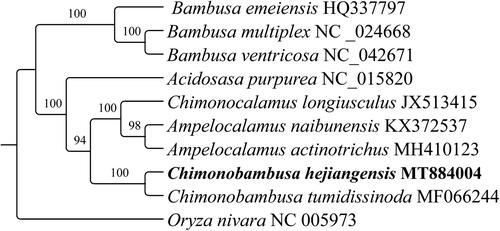
Phylogenetic analysis of ten complete cp genome sequences were selected to construct the phylogenetic tree using RAxML v.8.2.8 (Stamatakis Citation2014), and the bootstrap replicates parameter was set to 1,000. Oryza nivara was selected as an outgroup. The maximum likelihood (ML) tree showed that the genus Bambusa constitutes an isolated evolutionary branch, becoming a monophyletic group, a conclusion that is consistent with previous report (Zhang et al. Citation2019). Also, the reconstructed phylogeny revealed that C. hejiangensis is sister to C. tumidissinoda with strong support (100%) within the Chimonobambusa genus (). This cp genome can provide a theoretical basis for the further study of the Bambusoideae.
Acknowledgements
The authors thank other members of the laboratory for their help and suggestions for this research.
Disclosure statement
The authors declare that there are no conflicts of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MT884004.1/) under the accession MT884004. The associated BioProject, SRA, and Bio-Sample number are PRJNA689348, SRR13346655 and SAMN17207071 respectively.
References
- Ceccherini M, Poté J, Kay E, Van VT, Maréchal J, Pietramellara G, Nannipieri P, Vogel TM, Simonet P. 2003. Degradation and transformability of DNA from transgenic leaves. Appl Environ Microbiol. 69(1):673–678.
- Gou GQ, Ding YL, Yang L, Li L, Jiang WX. 2010. Nutrient analysis on bamboo shoots of three species in Chimonobambusa. China Veg. 16:79–81.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu HL, Wu M, Li F, Gao YM, Chen F, Xiang Y. 2018. TCP transcription factors in moso bamboo (Phyllostachys edulis): genome-wide identification and expression analysis. Front Plant Sci. 9:1263.
- Liu YJ, Wang LP, Fu B, et al. 2012. Nutrition components in shoots of different provenance of Chimonobambusa hejiangensis and C. pachystachys. Zhejiang Sci Technol. 32(04):37–42.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wambugu PW, Brozynska M, Furtado A, Waters DL, Henry RJ. 2015. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci Rep. 5:13957.
- Wu F-H, Kan D-P, Lee S-B, Daniell H, Lee Y-W, Lin C-C, Lin N-S, Lin C-S. 2009. Complete nucleotide sequence of Dendrocalamus latiflorus and Bambusa oldhamii chloroplast genomes. Tree Physiol. 29(6):847–856.
- Xue S, Shi T, Luo W, Ni X, Iqbal S, Ni Z, Huang X, Yao D, Shen Z, Gao Z, et al. 2019. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic Res. 6:89.
- Zhang X, Chen SY, Chen P, Liang H. 2019. The complete chloroplast genome of Chusquea culeou (Poaceae: Bambusoideae: Bambuseae). Mitochondrial DNA B. 4(1):91–92.
- Zhang YJ, Ma PF, Li DZ. 2011. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS One. 6(5):e20596.