Abstract
Tripsacum laxum (Guatemalan grass) is a perennial fodder grasses, which is commonly growing in large parts of Africa for a source of livestock feed. It has a high economic value as a forage. In this study, we obtained a complete chloroplast genome of T. laxum by Illumina sequencing. The results showed a circular genome of 140,556 bp, including the large single copy region (LSC, 82,939 bp), the small single-copy region (SSC, 12,573 bp), and a pair of 22,522 bp inverted repeat regions (IRs). The circular genome contained 120 genes, including 74 protein-coding genes, eight ribosomal RNA genes and 38 tRNA genes. Evolutionary relationship analysis indicates that T. laxum is more closely related to previously reported T. dactyloides.
Tripsacum laxum (Guatemalan grass) is a perennial fodder grasses, which belongs to the genus Tripsacum in Gramineae (http://www.iplant.cn/info/Tripsacum). This species is commonly growing in large parts of Africa for a source of livestock feed (Mwakha Citation1972; Topps Citation1986). T. laxum with a good adaptation can be tolerant in drought and acid, that is widely introduced as forage in other tropical regions of the world (Compere Citation1960; Paul et al. Citation2016). In recent years, T. laxum has also been cultivated in Southern China. Previous studies have demonstrated that it is effective for reducing run-off and soil loss (Madhu et al. Citation2011). The grass has important research significance and application value in the future. Hence, T. laxum is need to be better studied and prioritized as a considerable exploitation goal. In this study, we reported the chloroplast (cp) genome of T. laxum, which provided important genetic sources for molecular studying.
The fresh leaves were collected from Fujian Agriculture and Forestry University (Fuzhou, China; 26°5′16.04″N, 119°14′12.93″E). The voucher specimen of T. laxum was deposited in the National Engineering Research Center of Juncao Technology, Fujian Agriculture and Forestry University with specimen code FJ0623-TL01. The cp complete genomic DNA was extracted using a DNasecure Plant Kit (DP350-03) (TIANGEN, China). Therefore, we used the Illumina high-throughput sequencing to obtain the cp complete genome of T. laxum. The paired-end clean data were assembled by GetOrganelle kit (version 1.7.2) (Jin et al. Citation2020). The Geneious R21.0.1 (http://www.geneious.com/, Kearse et al. Citation2012) software was adopted to annotate the cp genome, while T. dactyloides (NC 037087.1) as reference sequences. Then, submitted to GenBank (accession number: MW387499).
The chloroplast sequence of T. laxum was 140,556 bp, including the large single-copy region (LSC, 82,939 bp), the small single-copy region (SSC, 12,573 bp), and a pair of 22,522 bp inverted repeat regions (IRs). The circular genome contained 120 genes, including 74 protein-coding genes (68 PCG species), eight ribosomal RNA genes (4 rRNA species) and 38 tRNA genes (21 tRNA species). The overall nucleotide composition of chloroplast genome is: 43,333 bp A (30.8%), 43,210 bp T (30.7%), 26,929 bp C (19.2%), 27,084 bp G (19.3%), and the total GC content of 38.4%.
The sequences alignments with MAFFT v7.304 (Katoh and Standley Citation2013) and MEGAX (Kumar et al. Citation2018) software was used to construct a maximum likelihood (ML) tree with 1000 bootstrap replications. The ML method based on 15 published complete chloroplast genome sequences which were downloaded from NCBI GenBank database, and constructed a phylogenetic tree () to confirm the relationship between T. laxum and other related taxa, while Oryza rufipogon, Oryza sativa were used as outgroups.
Figure 1. The maximum likelihood (ML) phylogenetic tree based on 15 complete chloroplast genomes, and Oryza rufipogon, Oryza sativa were used as outgroups.
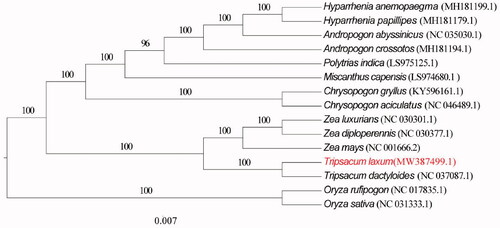
Phylogenomic analysis was conducted by RAxML v.8.2.8 (Stamatakis Citation2014). The ML phylogenetic analysis showed that T. laxum is closely related to T. dactyloides (NC 037087.1) in Gramineae ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings at this study have been stored in the NCBI database under accession number of MW387499 (https://www.ncbi.nlm.nih.gov/) and the NCBI SRA (BioProject: PRJNA688126) accession number of SRR13311453 (https://www.ncbi.nlm.nih.gov/sra).
Additional information
Funding
References
- Compere R. 1960. Introduction into Kivu (Congo) of T. laxum, a forage plant for dairy cows in the dry season. Bulletin Agricole du Congo Belge. 51(5):1085–1103.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Madhu M, Raghunath B, Tripathi KP, Sam MJ, Chandran B, Mohanraj R, Haldorai B. 2011. Vegetative barrier with contour staggered trenches for resource conservation in new tea plantations of the Nilgiris. Indian J Soil Conserv. 39(1):33–36.
- Mwakha E. 1972. Effect of cutting frequency on productivity of Napier and Guatemala grasses in Western Kenya. East African Agri Forest J. 37(3):206–210.
- Paul BK, Muhimuzi FL, Bacigale SB, Benjamin MMW, Chiuri WL, Amzati GS, Maass BL. 2016. Towards an assessment of on-farm niches for improved forages in Sud-Kivu, DR Congo. J Agri Rural Deve Tropics Subtropics. 117(2):243–254.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Topps JH. 1986. Pasture improvement research in Eastern and Southern Africa: Proceedings of a Workshop held in Harare, Zimbabwe, 17–21 September, 1984. Edited by Jackson A. Kategile. Intl Development Res. Centre, Ottawa, Canada. Agric Wastes. 17(2):157–158.
