Abstract
The complete mitochondrial genome of the Haifa grouper, Hyporthodus haifensis (Ben-Tuvia, 1953), has been obtained, through Illumina next-generation sequencing, and annotated. This mitogenome was found to be 16,525 bp long and to contain 37 genes, a control region, and the L-strand replication origin. The overall base composition of the complete mitogenome for this species was found to be 28.55% A, 28.07% C, 16.32% G, and 27.06% T. This study also looked into the mitogenome phylogenetic relationships of H. haifensis within the tribe Epinephelini and adds to the genetic resources currently available for the species.
The Haifa grouper, Hyporthodus haifensis (Ben-Tuvia, 1953), belongs to the family Serranidae, subfamily Epinephelinae, tribe Epinephelini. The latter is composed of 16 genera and 171 species, with the genus Hyporthodus hosting 18 species (Heemstra and Randall Citation1993; Parenti and Randall Citation2020). Epinephelini species are renowned for their economic value and form an integral part of coastal fisheries also in the Mediterranean Sea (Vella and Vella Citation2016). The life-history of most Epinephelini makes these species vulnerable to overfishing (Sadovy Citation1994; Sadovy de Mitcheson et al. Citation2013; Giglio et al. Citation2017), with a number of them being listed in high-risk categories by IUCN (Citation2020). Some Epinephelini species, such as Epinephelus marginatus, are considered as flagship species (Buchholz-Sørensen and Vella Citation2016) thus frequently studied, while others, such as H. haifensis, are rarely considered and are occasionally misidentified at fisheries landing sites with other groupers, such as Mycteroperca rubra, E. marginatus and Epinephelus caninus or with other species from unrelated taxa, such as Polyprion americanum (Vella and Vella Citation2016; pers. obs.). The latest IUCN assessment listed H. haifensis as Least Concern from its previous Data Deficient status (Francour and Pollard Citation2018), though the same report indicates that the species is naturally rare and due to the lack of life-history information should be closely monitored.
A 14.8 kg H. haifensis specimen was caught in February 2017 by local artisanal fishermen off the coast of Malta (35°45′N, 14°17′E). A tissue sample was collected from this specimen and deposited at the Ichthyological Collection of the Conservation Biology Research Group, University of Malta (www.um.edu.mt, Adriana Vella, [email protected]) under the voucher number CBRG 170202019. The genomic DNA was extracted from the tissue sample using GF-1 DNA Extraction Kit (Vivantis Technologies, Subang Jaya, Malaysia), and a DNA library of the whole genome was constructed. Next-generation sequencing was used to sequence 2 × 150 bp end reads through Illumina HiSeqX (Illumina, San Diego, CA). Sequences were paired, trimmed at Q ≥ 30 and the complete circular mitogenome was de novo assembled using Geneious R10 (Kearse et al. Citation2012). The tRNA genes were identified through secondary structures using tRNAscan-SE version 2.0 (Chan and Lowe Citation2019), while protein-coding genes (PCGs), rRNA genes and the control region were identified through homology with other Epinephlini species (Zhuang et al. Citation2013).
The complete mitogenome for H. haifensis is 16,525 bp long (MW015093), falling in the range of other mitogenomes of Epinephelini species, which range between 16,389 bp in the Striped grouper, Epinephelus latifasciatus (KC480177, Lai et al. Citation2013) to 17,227 bp in the Duskytail grouper, Epinephelus bleekeri (KF556648, Wu et al. Citation2015). The mitogenome studied here contains 13 PCGs, two rRNA genes, 22 tRNA genes, and two non-coding regions (control region and OL) and follows the typical gene order of fish species (Satoh et al. Citation2016). The majority of the genes are encoded on the H-strand, except ND6 gene, eight tRNA genes, and OL. The PCGs lengths range between 168 bp (ATP8) and 1839 bp (ND5). Most of the PCGs utilize ATG as their start codon except COX1 which uses GTG similar to most other fish species (Satoh et al. Citation2016), and ATP6 which uses CTG. In general, the latter start codon for ATP6 is considered as unusual in fish species (Satoh et al. Citation2016), however, it was found to occur in 82% of the Epinephelini species included in . Almost all genes have TAA as their stop codon, except ND3 that uses TAG, while COX2 and ND4 use the incomplete stop codon T–. The length of the 22 tRNA genes varies between 67 bp (Cys) to 77 bp (LeuUUR), and all produced the expected cloverleaf structure except for SerAGY that has the DHU arm missing.
Figure 1. Bayesian Inference based phylogeny depicting the mitogenomic relationship (excluding the control region) between 33 Epinephelini species using other Serranidae species as outgroup. Each label includes the GenBank accession number, species, and reference (listed in the Supplementary Material). The numbers at the nodes indicate the posterior probability values. This analysis used 5 × 106 generations, a sample frequency every 1,000 generations and a burn-in of 25%.
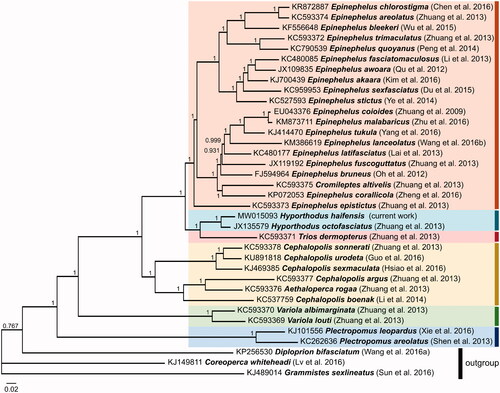
The mitogenome of H. haifensis was aligned with that of other Epinephelini species using ClustalW (Thompson et al. Citation1994), and a phylogenetic tree was constructed using Bayesian Inference analysis through MrBayes version 3.2.6 (Huelsenbeck and Ronquist Citation2001) () using GTR G + I substitution model as determined by jModelTest version 2.1.7 (Darriba et al. Citation2012). This analysis placed H. haifensis on the same branch as Hyporthodus octofasciatus, forming a sister group to Triso dermopterus. All species grouped together according to their respective genera, except for the monotypic genus Cromileptes (represented by C. altivelis) which nested within the genus Epinephelus and the monotypic genus Aethaloperca (represented by A. rogaa) which nested within the genus Cephalopholis. The position of these two monotypic genera is consistent with observations noted using other genetic markers (Craig and Hastings Citation2007) and other smaller data sets of Epinephelini mitogenomes (Zhuang et al. Citation2013). This outcome further supports the need for a taxonomic revision of these monotypic taxa within their respective clades as indicated in Zhuang et al. (Citation2013). This study adds to the genetic resources available for H. haifensis, which can be used as a tool to promote further research and effective conservation of this rare species.
Ethical approval
This study did not require ethical approval as it made use of muscle tissue collected from a dead specimen that was caught by a local fisherman and was sold at the local fish market.
Supplemental Material
Download MS Word (14.8 KB)Acknowledgments
The authors would like to thank Maltese fishermen and the Ministry for Sustainable Development, the Environment, and Climate Change for supporting this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MW015093. The associated BioProject number is PRJNA661720.
Additional information
Funding
References
- Buchholz-Sørensen M, Vella A. 2016. Population structure, genetic diversity, effective population size, demographic history and regional connectivity patterns of the endangered Dusky grouper, Epinephelus marginatus (Teleostei: Serranidae), within Malta’s Fisheries Management Zone. PLOS One. 11(7):e0159864.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Craig MT, Hastings PA. 2007. A molecular phylogeny of the groupers of the subfamily Epinephelinae (Serranidae) with a revised classification of the Epinephelini. Ichthyol Res. 54(1):1–17.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Francour P, Pollard DA. 2018. Hyporthodus haifensis. The IUCN red list of threatened species 2018: e.T132828A100570068.
- Giglio VJ, Bender MG, Zapelini C, Ferreira CEL. 2017. The end of the line? Rapid depletion of a large-sized grouper through spearfishing in a subtropical marginal reef. Perspect Ecol Conserv. 15(2):115–118.
- Heemstra PC, Randall JE. 1993. Groupers of the world. FAO fisheries synopsis no. 125, vol 16., Rome: FAO.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- IUCN. 2020. The IUCN red list of threatened species. Version 2020-2. https://www.iucnredlist.org.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lai T, He B, Peng Z, Wang X, Pan L. 2013. Complete mitochondrial genome of the striped grouper Epinephelus latifasciatus (Serranidae, Epinephelinae). Mitochondrial DNA. 24(5):510–512.
- Parenti P, Randall JE. 2020. An annotated checklist of the fishes of the family Serranidae of the world with description of two new related families of fishes. FishTaxa. 15:1–170.
- Sadovy de Mitcheson Y, Craig MT, Bertoncini AA, Carpenter KE, Cheung WL, Choat JH, Cornish AS, Fennessy ST, Ferreira BP, Heemstra PC, et al. 2013. Fishing groupers towards extinction: a global assessment of threats and extinction risks in a billion dollar fishery. Fish Fish. 14(2):119–136.
- Sadovy YJ. 1994. Grouper stocks of the western central Atlantic: the need for management and management needs. Ann Gulf Caribb Fish Inst Proc. 43:43–64.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 17:719.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680.
- Vella A, Vella N. 2016. Genetic barcoding and preliminary phylogenetic analysis of Serranidae species from Maltese coastal waters, with a perspective on their Mediterranean phylogeography. Nat Eng Sci. 1(3):66–77.
- Wu X, Xie Z, Yang L, Yang H, Yue L, Hou L, Zhang Y, Shu H. 2015. The complete mitochondrial genome of the duskytail grouper Epinephelus bleekeri (Serranidae: Epinephelinae). Mitochondrial DNA. 26(5):722–723.
- Zhuang X, Qu M, Zhang X, Ding S. 2013. A comprehensive description and evolutionary analysis of 22 grouper (Perciformes, Epinephelidae) mitochondrial genomes with emphasis on two novel genome organizations. PLoS One. 8(8):e73561.
