Abstract
The complete mitochondrial genome of Sinularia penghuensis was sequenced and analyzed using next-generation sequencing. The present mitochondrial genome was 18730 bp in length, containing 14 protein-coding genes (PCGs) (cox1-cox3.nad1-nad6, nad4L, atp6, atp8, cytb, and MutS), two ribosomal RNA genes (rRNAs) (12S and 16S), and one transfer RNA gene (Met-tRNA). The phylogenetic analysis of family Alcyoniidae revealed that S. penghuensis and Sinularia maxima cluster together. Five species in Sinularia reveals high identity in mitogenome sequences that the lowest variable sites (SNPs) were found between S. penghuensis and S. maxima.
Coral ecosystem is not only the largest ecosystem in the ocean, but it is the most complex aquatic ecosystem. Coral reefs provide the highest biodiversity in all marine ecosystems (Baums Citation2008; Keller et al. Citation2017). Genus Sinularia is the largest genus of the family Alcyoniidae with approximately 170 species. Sinularia contain compounds with unique structure and bioactivity, which makes it an important natural resource (Lakshmi and Kumar Citation2009). Sinularia penghuensis is usually in greenish brown or turquoise gray color, grows on the reef platform or slope with a depth of 3–15 m, and it also distributed in the sea area with slightly turbid water quality.
In this study, the complete mitochondrial genome (mitogenome) of S. penghuensis Ofwegen and Benayahu, 2012 (GenBank: MW256412) was sequenced using next-generation sequencing. The specimen of S. penghuensis was collected from the coral reef in West Island (Sanya, Hainan province, China; 18° 14′ 5.93′′ N, 109° 22′ 46.46′′ E), and deposited at Hainan Tropical Ocean University Museum of Zoology (www.hntou.edu.cn, Pei-Zheng Wang, [email protected]) under the voucher number NO. 0007-Sp). The identification of the specimen was performed using PuCAs-mtMutS and PuCAs-28S. The total genomic DNA was extracted and used for sequencing on Illumina HiSeq 4000 platform (Novogene: Tianjin, China).
10G raw data was obtained followed by de novo assembling and mapping to the reference Sinularia mitochondrial genome (Sinularia ceramensis, NC_044122) with Spades version 3.9.0 (Bankevich et al. Citation2012) and bowtie version 2.2.9 (Langmead and Salzberg Citation2012). Protein-coding genes (PCGs) and ribosomal RNA genes (rRNAs) were annotated using alignment with a large number of Alcyonacea mitogenome available in genbank and the online gene detection server NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Position and secondary structure of transfer RNA gene (tRNA) was performed by online server tRNAscan-SE2.0 (http://lowelab.ucsc.edu/tRNAscan-SE/).
The complete mitogenome of S. penghuensis was 18,730 bp in size, with a base composition of 30.19% A, 34.00% T, 19.34% G, and 16.47% C. The base composition of the complete mitogenome reveals strong AT bias, the AT content of whole mitogenome was 64.19%, and AT content of PCGs, tRNA, and rRNAs were 65.50%, 54.93%, and 56.95%, respectively.
Totally 17 genes were detected on the mitogenome, including 14 PCGs, two rRNAs, and one tRNA. Four PCGs (cox2, cox3, atp6, and atp8) and the only tRNA gene (tRNA-Met) were located on the light strand and other genes (cox1, cytb, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, mutS, 12S, and 16S) were encoded on the heavy strand. This species has identical gene order with the other published mitogenome in Sinularia.
ATG was the only selected start codon among all PCGs. Eight genes (nad1, cytb, nad6, nad3, nad2, nad5, cox3, and cox2) were stopped by TAG, and five genes (nad4L, mutS, nad4, atp6, and atp8) use TAA as stop codon. Cox1 was stopped by premature stop codon T, which was presumably to form TAA codon via posttranscriptional polyadenylation (Boore JL Citation2001). Only one tRNA, tRNA-Met, was detected that can be folded into typical clover-leaf secondary structures. Totally 15 intergenic regions were existed, which range from 4 to 112 nucleotides.
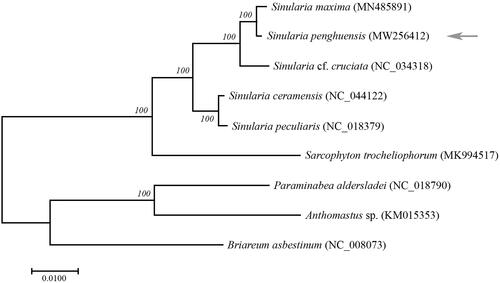
The new present mitogenome and all sequences mitogenomes in Alcyoniidae were used in phylogenetic analysis (Brockman and Mcfadden Citation2012; Kayal et al. Citation2013; CitationFigueroa and Baco 2015; Shimpi et al. Citation2017; Asem et al. Citation2019; Chen et al. Citation2019; Shen et al. Citation2019), and Briareum asbestinum (Medina et al. Citation2006) was set for the outgroup. The concatenated dataset for nucleotides including all 14 PCGS and two rRNAs were performed to draw phylogenetic tree using the maximum-likelihood method (ML). The generate phylogenetic tree topology revealed that the species of genus Sinularia cluster together and divided with the Sarcophyton, and S. penghuensis has the most closed relationship with Sinularia maxima (). The clade of (S. penghuensis+ S. maxima) + Sinularia cf. cruciata was sister with the other two Sinularia species (Sinularia peculiaris + Sinularia ceramensis). The sequences of mitogenomes in Sinularia species exhibited a high degree of consistency. The variable sites (SNPs) between S. penghuensis and other Sinularia species (S. maxima, S. cruciate, S. ceramensis, and S. peculiaris) were 53, 189, 381, and 391, respectively. It could be considered that S. penghuensis and S. maxima are the most closed species, which was consistent with the result of phylogenic analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW256412.
Additional information
Funding
References
- Asem A, Lu H, Wang PZ, Li WD. 2019. The complete mitochondrial genome of Sinularia ceramensis Verseveldt, 1977 (Octocorallia: Alcyonacea) using next-generation sequencing. Mitochondrial DNA Part B. 4(1):815–816.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Baums IB. 2008. A restoration genetics guide for coral Reef conservation. Mol Ecol. 17(12):2796–2811.
- Boore JL. 2001. Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Mol Biol Evol. 18(7):1413–1416.
- Brockman SA, Mcfadden CS. 2012. The mitochondrial genome of Paraminabea aldersladei (cnidaria: Anthozoa: Octocorallia) supports intramolecular recombination as the primary mechanism of gene rearrangement in octocoral mitochondrial genomes. Genome Biol Evol. 4(9):994–1006.
- Chen Y, Dan Y-T, Lu H, Wang P-Z, Asem A, Li W. 2019. The complete mitochondrial genome of Sinularia maxima Verseveldt, 1971 (Octocorallia: Alcyonacea) using next-generation sequencing. Mitochondrial DNA Part B. 4(2):3425–3426.
- Figueroa DF, Baco AR. 2015. Octocoral mitochondrial genomes provide insights into the phylogenetic history of gene order rearrangements, order reversals, and Cnidarian phylogenetics. Gen Biol Evol. 7(1):391–409.
- Kayal E, Roure B, Philippe H, Collins AG, Lavrov DV. 2013. Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evol Biol. 13:5.
- Keller NB, Oskina NS, Savilova TA. 2017. Distribution of deep-water Scleractinian corals in the Atlantic Ocean. Oceanology. 57(2):298–305.
- Lakshmi V, Kumar R. 2009. Metabolites from Sinularia species. Nat Prod Res. 23(9):801–850.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Medina M, Collins A, Takaoka T, Kuehl J, Boore J. 2006. Naked corals: skeleton loss in Scleractinia. Proc Natl Acad Sci USA. 103(24):9096–9100.
- Shen CY, Dan YT, Asem A, Wang PZ, Xue W, T XB, Li WD. 2019. The complete mitochondrial genome of soft coral Sarcophyton trocheliophorum (Cnidaria: Anthozoa) using next-generation sequencing. Mitochondrial DNA Part B. 4(2):3734–3735.
- Shimpi GG, Vargas S, Poliseno A, Worheide G. 2017. Mitochondrial RNA processing in absence of tRNA punctuations in octocorals. BMC Mol Biol. 18(1):16.