Abstract
The complete chloroplast genome of Viola philippica was sequenced, assembled, and annotated. It is a circular form of 156,469 bp in length, which was separated into four distinct regions, a large single-copy (LSC) of 85,668 bp, a small single-copy region (SSC) of 18,001 bp, and two inverted repeats (IR) of 26,400 bp. After annotation, a total of 129 genes were predicted, of which, 84 encode proteins, 8 rRNA, and 37 tRNA. The evolutionary history, inferred using maximum likelihood (ML) method, indicates that V. philippica was grouped within Violaceae, and comprised a clade with Viola seoulensis with 100% Bootstrap value.
Viola philippica, belonging to Violaceae, is a widely distributed perennial herb in China (Zhang et al. Citation2014). For containing bioactive constituents, such as Lignans, flavonoids, and coumarins, it was used extensively in traditional Chinese medicines for heat-clearing, detoxification, anti-inflammation, and pain relief (He et al. Citation2011; Wang et al. Citation2019; Yu et al. Citation2019). In previous studies, although many literatures on chemical composition and their functions have been documented, only a few literatures were about its genetic research (Zhang et al. Citation2014; Li et al. Citation2016). In this study, we report the complete chloroplast (cp) genome of V. philippica.
Samples from Qilian mountains (36°35′18″N, 101°49′33″E) in Qinghai province were collected for sequencing. Voucher specimen (HCPQNU-20200602001) was deposited in the Herbarium, College of Pharmacy, Qinghai Nationalities University. A sample’s total genomic DNA was extracted from about 100 mg fresh leaves using a modified CTAB method (Murray and Thompson Citation1980). Paired-end Libraries with an average length of 350 bp were constructed and sequenced on Illumina Novaseq 6000 platform (Shenzhen Huitong Biotechnology Co. Ltd, Shenzhen, China). The complete cp genome was assembled via the de novo assembler SPAdes (Bankevich et al. Citation2012) and annotated via PGA (Qu et al. Citation2019) with Viola seoulensis (KP749924) chloroplast genome as reference genome.
The complete cp genome of V. philippica (GenBank accession no. MT796627.1) has a typical quadripartite form of 156,469 bp in length, and composed of a large single-copy region (LSC, 85,668 bp), a small single-copy region (SSC, 18,001 bp), and two inverted repeats (IR, 26,400 bp). GC content of the genome is 36.3%. A total of 129 genes were predicted on this cp genome, of which, 84 encode proteins, 8 rRNA, and 37 tRNA.
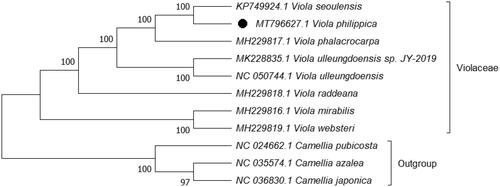
Phylogenetic analysis was performed based on complete cp genomes of V. philippica and other seven related species reported in Violaceae, three species in Theaceae as out-group. The sequences were aligned using HomBlocks (Bi et al. Citation2018). The evolutionary history was inferred using maximum likelihood (ML) method in MEGA X (Kumar et al. Citation2018) with general time-reversible nucleotide substitution, Gamma distributed (GTR + G) model, and partial deletion of gaps/missing data. Bootstrap (BS) values were calculated from 1000 replicate analysis (). As expected, V. philippica was grouped within Violaceae, and comprised a clade with Viola seoulensis with 100% BS value. The complete cp genome of V. philippica will be helpful for further studies on population genetics, taxonomy, or resource protection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MT796627.1) under the accession no. MT796627.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA678799, SRR13070133, and SAMN16812861, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bi GQ, Mao YX, Xing QK, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- He WJ, Chan LY, Zeng GZ, Daly NL, Craik DJ, Tan NH. 2011. Isolation and characterization of cytotoxic cyclotides from Viola philippica. Peptides. 32(8):1719–1723.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li QX, Huo QD, Wang J, Zhao J, Sun K, He CY. 2016. Expression of B-class MADS-box genes in response to variations in photoperiod is associated with chasmogamous and cleistogamous flower development in Viola philippica. BMC Plant Biol. 16(1):151.
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8(19):4321–4326.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Wang YL, Zhang L, Li MY, Wang LW, Ma CM. 2019. Lignans, flavonoids and coumarins from Viola philippica and their α-glucosidase and HCV protease inhibitory activities. Nat Prod Res. 33(11):1550–1555. ]
- Yu Q, Liu MZ, Xiao HH, Wu ST, Qin XL, Lu ZJ, Shi DQ, Li SQ, Mi HZ, Wang YB, et al. 2019. The inhibitory activities and antiviral mechanism of Viola philippica aqueous extracts against grouper iridovirus infection in vitro and in vivo. J Fish Dis. 42(6):859–868.
- Zhang YJ, Liu H, Zhang XH, Wang SR, Liu CX, Yu C, Wang XJ, Xiang WS. 2014. Micromonospora violae sp. nov., isolated from a root of Viola philippica Car. Antonie Van Leeuwenhoek. 106(2):219–225.