Abstract
Paraqianlabeo lineatus is a small-sized fish which is endemic to Guizhou province, China. The complete mitochondrial genome of P. lineatus is 16,598 bp in total length, with 37 genes, including 13 PCGs, 22 tRNA genes, two rRNA genes (16S and 12S) and a non-coding region (D-loop). The positions and sequences of genes were consistent with congeners of Labeoninae. The nucleotide composition of the mitogenome was A (31.5%), T (26.7%), G (15.9%), C (25.8%) and was slightly A + T biased. Phylogenetic analysis conducted using Bayesian Inference method showed that P. lineatus clustered with Pseudogyrincheilus procheilus within the subfamily Labeoninae. The results may provide helpful data for further studies of the evolutionary history of Labeoninae.
Paraqianlabeo lineatus is a small-sized fish which is endemic to drainages of Yangtze river, Chishui river and Wu-Jiang river, Guizhou province, China (Zhao et al. Citation2014). This monotypic species is usually found in upper hill streams with rocks. There are few ecological and molecular data about P. lineatus (Li et al. Citation2016; Zhao Citation2016). Here, we provide the first description of the complete mitochondrial genome of P. lineatus. The sequence was obtained from a specimen caught in Mengxi river, Shizhi village, Zhengan county (E 107°17′37″, N 28°17′17″), Guizhou province, a tributary flowing into Wu-Jiang river. Total genomic DNA was extracted from the pelvic fin preserved in 95% alcohol using the Qiagen QIAamp tissue kit following the manufacturer’s protocol. The complete mitochondrial genome was sequenced by next-generation sequencing methods and the annotated sequence was archived at Genebank (accession number MW039085). The specimen (GZ-20170217, 7.7 cm in standard length, 5.0 g in weight) was deposited in the collection of fisheries research institution, Guizhou Academy of Agricultural Sciences.
Structure of the mitogenome of P. lineatus was consistent with those of other fishes (Zheng and Yang Citation2016; Tan et al. Citation2019; Pan et al. Citation2020), consisting 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and a non-coding regions. Most of the coding regions are encoded on the H-strand except for ND6, and there are eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu and tRNAPro) encoded on H-strand. The base composition of protein-coding genes showed a weak anti-G bias (15.3%) whereas the anti-G bias was strongly observed in the third codon position (6.7%), overall base composition of A + T content (58.2%) is nearly the same with coding region genes (58.8%). Almost all of the 13 protein-coding genes started with the typical start codon ATG but COI, which started with GTG. For the stop codon, six of them share the complete stop codon, while seven shared the incomplete stop codon with a terminal T or TA. The D-Loop was located between tRNAPho and tRNAPhe with 936 bp, the base composition reflected a strong A + T-rich (65.7%), richer than the overall average content (58.2%).
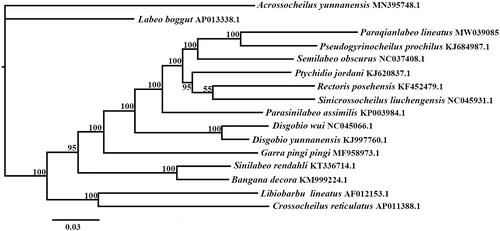
We chose Bayesian Inference method to infer evolutionary relationships based on complete mitogenome between P. lineatus and 15 other sequences downloaded from Genebank, Acrossocheilus yunnanensis (Cyprinidae: Barbinae) was chosen as outgroup here. The phylogenetic tree showed that P. lineatus clustered with Pseudogyrincheilus procheilus with high Posterior Probability (), consistent with previous studies based on short mitochondrial DNA sequences (Zhao Citation2016; Zheng et al. Citation2016). this may tell us that genus Paraqianlabeo and Pseudogyrincheilus have more closely evolutionary relationship, and the results would provide references for further study of subfamily Labeoninae.
Acknowledgements
Our sincere thanks are given to Wei Wang and Wen-Wu Min for helping with field work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW039085. The associated BioProject, Bio-Sample and SRA numbers are PRJNA694796, SAMN17574689 and SRR13664247 respectively.
Additional information
Funding
References
- Li Y, Zhao H, Peng ZG, Zhang Y. 2016. Isolation and characterization of 21 polymorphic microsatellite markers for a new Labeonine fish (Paraqianlabeo lineatus zhao et al. 2014) using Illumina paired-end sequencing. J Appl Ichthyol. 32(1):126–128.
- Pan XY, Shao LY, Lao JQ, Kuang TX, Chen JC, Zhou L. 2020. Mitochondrial genome of Parasinilabeo longicorpus provides insights into the phylogeny of Labeoninae. Mitochondr DNA B. 5(3):3062–3063.
- Tan HY, Zhang M, Cheng GP, Chen XL. 2019. The complete mitochondrial genome of Discogobio laticeps (Cyprinidae: Labeoninae). Mitochondr DNA B. 4(1):1981–1982.
- Zhao HT, Sullivan JP, Zhang YG, Peng ZG. 2014. Paraqianlabeo lineatus, a new genus and species of Labeonine fishes (Teleostei: Cyprinidae) from south china. Zootaxa. 3841(2):257–270.
- Zhao HT. 2016. The genetic diversity and population structure of a new genus and species of Labeonine fishes. Chongqing: Southwest University.
- Zheng LP, Chen XY, Yang JX. 2016. Molecular systematics of the Labeonini inhabiting the karst regions in southwest China (Teleostei, Cypriniformes). ZK. 612:133–148.
- Zheng LP, Yang JX. 2016. Complete mitochondrial genome of two endemic species to China: Discocheilus wui and Discocheilus wuluoheensis (Cypriniformes, Cyprinidae). Mitochondrial DNA B Resour. 1(1):851–852.