Abstract
Viburnum burejaeticum Regel et Herd is widely cultivated in botanical gardens. However, as a member of Adoxaceae, few studies have been carried out on its phylogenetic relationship with other family members. Here we report the first complete chloroplast (cp) genome sequence of V. burejaeticum collected from China. The circular cp genome is 158,381 bp in size, including a large single-copy (LSC) region of 87,067 bp and a small single-copy (SSC) region of 18,212 bp, which were separated by two inverted repeat (IR) regions (26,551 bp each). A total of 126 genes were annotated, including 8 ribosomal RNAs (rRNAs), 36 transfer RNAs (tRNAs), and 82 protein-coding genes (PCGs). The sequence comparison of two V. burejaeticum collected from Korea and China revealed 101 single nucleotide polymorphisms (SNPs) and 16 insertions/deletions (InDels). In addition, maximum-likelihood (ML) phylogenetic analysis indicated V. burejaeticum species collected in Korea and China are clustered together. This study provides useful information for future genetic study of V. burejaeticum.
Viburnum burejaeticum Regel et Herd (Adoxaceae family) is widely distributed in China, Korea, and Europe. Because of its high ornamental value, V. burejaeticum is commonly cultivated in botanical gardens. The active ingredients of viburnum plants are mainly polyphenols, polysaccharides, and steroids (Wang et al. Citation2010; Shao et al. Citation2019). The leaves of V. burejaeticum can be used as medicine to treat scabies and Arthritic diseases. There are many researches on the phylogenetic relationship of genus Viburnum (Clement et al. Citation2014). However, few genetic and genomic studies have been done on V. burejaeticum. Recently, chloroplast genome sequence of V. burejaeticum collected from Korea has been reported (GenBank accession number: NC_050741). In this work, the complete chloroplast (cp) genome of V. burejaeticum collected from China has been reported.
The young and healthy leaf samples were collected from School of Traditional Chinese medicine, Jilin Agriculture Science and Technology College, Jilin, Jilin Province, China (44°3′5.44″N, 126°6′34.44″E, 237 m above sea level). The voucher specimen (accession number: GL202003001) was laid in the herbarium of the School of Life Science of Guizhou Normal University (Jinghua Mu, [email protected]). The total DNA (accession number: GL202003002) extracted using DNAsecure Plant Kit (TIANGEN, Beijing, China) was stored at −80 °C refrigerator of the laboratory (room number: 1403) of School of Life Science, Guizhou Normal University. Qualified DNA (700 ng) was used for library construction. Sequencing libraries were generated using NEB Next® Ultra DNA Library Prep Kit for Illumina® (NEB, Ipswich, MA). The library preparations were sequenced on an Illumina platform and 150 bp paired-end reads were generated. The filtered reads were assembled using the program GetOrganelle (Jin et al. Citation2019). The assembled cp genome was annotated using the online software GeSeq (Tillich et al. Citation2017). The complete cp genome was submitted to GenBank under the accession number MW296956.
The length of the complete cp genome sequence of V. burejaeticum (China collection) is 158,381 bp, which is sorted into four sections, consisting of a large single-copy (LSC; 87,067 bp) region, a small single-copy (SSC; 18,212 bp) region, and two inverted repeat (IRA and IRB) regions of 26,551 bp each. A total of 126 genes were predicted in the cp genome of V. burejaeticum (China collection), including 82 protein-coding (PCGs), 8 ribosomal RNA (rRNA), and 36 transfer RNA (tRNA) genes. Among the assembled genes, all rRNAs, 6 PCGs (rps7, rps12, rpl2, rpl23, ndhB, and ycf2) and 8 tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, and trnV-GAC) with double copies. Intron-exon analysis indicated 106 genes with no introns and 20 genes harbored introns.
The full-length cp genome of V. burejaeticum (China collection) was 2 bp shorter than Korea collection. We identified 101 single nucleotide polymorphisms (SNPs) and 16 insertions/deletions (InDels) between these two V. burejaeticum collections. This result indicated that these two samples have richer intra-species diversity, like some other species (Nguyen et al. Citation2017, Citation2020; Kim et al. Citation2018). The different environment maybe an important cause of this abundant intra-species diversity.
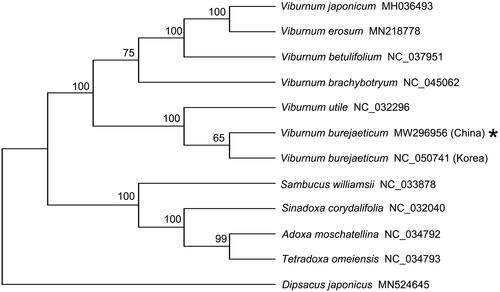
To further understand, the chloroplast genome of we assembled, the phylogenetic tree of China V. burejaeticum cp plastid genomes was constructed with closely related plants belonging to the Adoxaceae, including six species in Viburnum genus (containing Korea V. burejaeticum), one species in Sambucus genus, one specie in Sinadoxa genus, one specie in Adoxa genus, one specie in Tetradoxa genus, and one Caprifoliaceae member (Dipsacus japonicus) used as outgroup through maximum-likelihood (ML) analysis. The ML tree was performed using RAxML version 8.0.19 (Model: GTRGAMMA) (Stamatakis Citation2014) with 1000 bootstrap replicates. The phylogenetic tree clearly classified these 11 species into 2 distinct clusters (). In the Viburnum cluster, seven Viburnum species were grouped into a clade (). As expected, Korea and China collections of V. burejaeticum were clustered together ().
Figure 1. Maximum likelihood tree based on the complete cp genome sequences of 10 species from the Adoxaceae family shows the 2 distinct clusters. Dipsacus japonicus used as outgroup. GenBank accession numbers are described in the figure. Shown next to the nodes are bootstrap support values based on 1000 replicates.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The complete chloroplast genome data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession number MW296956. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682877, SRX9639380, and SAMN17013273, respectively.
Additional information
Funding
References
- Clement WL, Arakaki M, Sweeney PW, Edwards EJ, Donoghue MJ. 2014. A chloroplast tree for Viburnum (Adoxaceae) and its implications for phylogenetic classification and character evolution. Am J Bot. 101(6):1029–1049.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv.256479.
- Kim CK, Seol YJ, Perumal S, Lee J, Waminal NE, Jayakodi M, Lee SC, Jin S, Choi BS, Yu Y, et al. 2018. Re-exploration of U's triangle Brassica species based on chloroplast genomes and 45S nrDNA sequences. Sci Rep. 8(1):7353.
- Nguyen VB, Linh Giang VN, Waminal NE, Park HS, Kim NH, Jang W, Lee J, Yang TJ. 2020. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers. J Ginseng Res. 44(1):135–144.
- Nguyen VB, Park HS, Lee SC, Lee J, Park JY, Yang TJ. 2017. Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences. J Agric Food Chem. 65(30):6298–6306.
- Shao JH, Chen J, Xu XQ, Zhao CC, Dong ZL, Liu WY, Shen J. 2019. Chemical constituents and biological activities of Viburnum macrocephalum f. keteleeri. Nat Prod Res. 33(11):1612–1616.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang XY, Shi HM, Li XB. 2010. Chemical constituents of plants from the genus Viburnum. Chem Biodivers. 7(3):567–593.
