Abstract
In the study, the complete mitochondrial genome of Pyrops candelaria (Linnaeus) was determined for the first time. The genome was a circular molecule with 16,253 bp in full-length, consists of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and one control region. The nucleotide composition of Pyrops candelaria (Linnaeus) mitogenome was A: 47.13%, C: 15.9%, G: 10.79%, and T: 26.18%. Phylogenetic analysis showed that P. candelaria had a close relationship with to Lycorma delicatula.
Pyrops candelaria (Linnaeus) (Hemiptera: Fulgoridae) is a common pest in tropical and subtropical orchards, which is distributed in many countries in Southeast Asia (Wang et al. Citation2000). P. candelaria mainly damages many fruits such as longan, lychee, and kiwi (Zhang et al. Citation2015; Qian et al. Citation2018; Xu et al. Citation2019). Larvae and adults of P. candelaria suck sap from trunks, causing fruit tree death, leaf curl and withering, and serious yield loss, its excrement can induce sooty mold (Wang et al. Citation2000). The complete mitochondrial DNA (mtDNA) sequence of P. candelaria was determined which has not been reported before. This study will provide a reference for further studies on the taxonomy and genomics of P. candelaria.
The sample of P. candelaria was collected at Taxidermy Park in Guangxi University, Nanning, Guangxi Zhuang Autonomous Region, China (22°50′28.41″N, 108°17′9.00″E), and stored in 98% alcohol immediately. The specimen was deposited at the Herbarium of Guangxi University College of Agriculture (specimen code FDSW202318381-1r). The complete mitochondrial genome sequence has the accession number MW355618 in GenBank.
A total of 9.84 G of raw data and 9.42 G of clean data were obtained for sequence analysis by SPAdes (version 3.9) (Tamura and Nei Citation1993). The entire mitochondrial genome of P. candelaria was a closed 16,253 bp long circular molecule, and consists of 37 genes. These genes contained 13 protein-coding genes (ND2, COX1, COX2, ATP8, ATP6, COX3, ND3, ND5, ND4, ND4L, ND6, CYTB, and ND1), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and one control regions (D-loop). The AT content was 73.31% and the other bases were 47.13% A, 15.9% C, 10.79% G, and 26.18% T in the nucleotide composition of the mitotic genome of P. candelaria, that shows a great preference for AT.
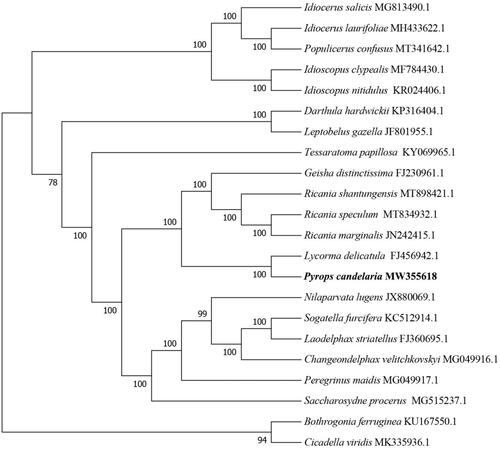
To investigate the phylogenetic relationships of P. candelaria in Fulgoridae, the phylogenetic tree was constructed by Neighbor-Joining method with 1000 bootstrap replicates, Molecular Evolutionary Genetics Analysis Version 7.0 (MEGA7.0) was used to make phylogenetic analysis (Kumar et al. Citation2016). The results showed that the mtDNA of P. candelaria was closely related to that of Lycorma delicatula ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW355618, accession number MW355618. The data are morally correct and does not violate the protection of human subjects or other valid ethical, privacy or security concerns.
Additional information
Funding
References
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Qian J, Huang D, Li D, Qian X, Wang Y. 2018. Investigation and symptom analysis of Imocarpus longana Lour pests in Wenchang city of Hainan province. Xiandai Hortic. 2:6–9.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10(3):512–526.
- Wang G, Huang J, Huang B. 2000. Studies on the biology of Fulgora candelaria (L.) (Homoptera: Fulgoridae). Entomol J East China. 9(1):61–65.
- Xu H, Quan L, Chen B, Dong Y. 2019. Analysis of pest community diversity and temporal dynamics in thinned and un-thinned litchi orchards. J Fruit Sci. 36(4):493–503.
- Zhang T, Yuan Y, Chang X, Teng H, Wang D. 2015. Insect diversity on Actinidia chinensis Planch in Shanghai. Acta Agric Shanghai. 31(6):49–52.